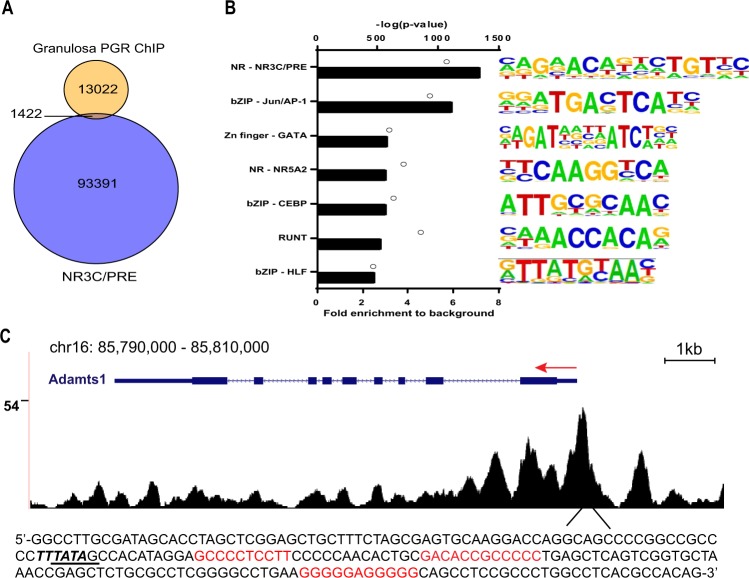
Figure 3.
PGR binding is enriched at PRE/NR3C in the genome, but PGR also associates with other transcription factor binding elements. (A) PGR binding sites in granulosa cells in relation to global PRE/NR3C locations. PRE/NR3C sites within the entire mouse genome were identified using FIMO in MEME Suite to search for the consensus PRE/NR3C sequence. Analysis of enriched motifs represented in granulosa cell PGR ChIP data showed that 10.51% of peaks overlapped with consensus PRE/NR3C sites. (B) Top most common known motifs found to be enriched at PGR binding sites in granulosa cells. Bars indicate fold enrichment of motif to background. Circles indicate –log(p-value). Among motifs found by HOMER that belonged to the same transcription factor family, the most conservative sequences were selected and ranked by fold enrichment compared to background frequency. (C) PGR binding sites in the mouse genome visualised through UCSC Genome Browser. The track shown is located in chromosome 16 at the genomic region for Adamts1. The red arrow indicates the TSS (arrow tail) and direction of transcription. The sequence of the most prominent PGR peak is listed below, with the TATA box underlined and previously described G/C-site in red.