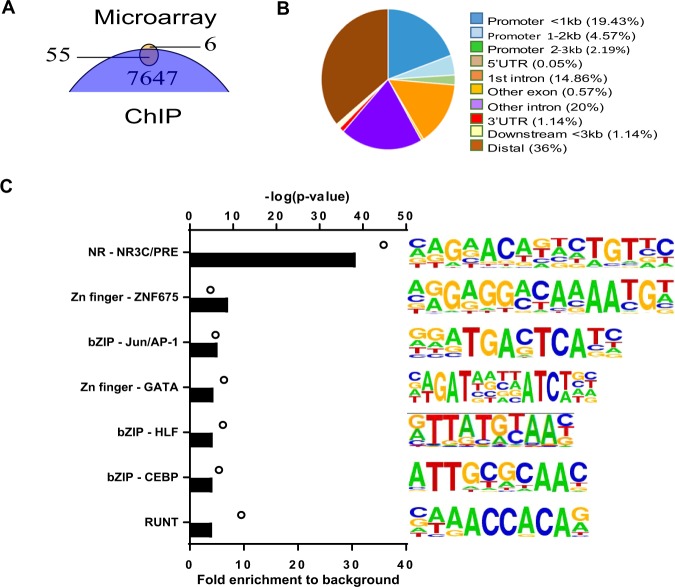
Figure 5.
Characteristic of PGR-dependent DEG with PGR binding in peri-ovulatory granulosa cells. (A) Venn diagram showing DEG in microarray in relation to genes with PGR binding sites from ChIP-seq (B) Genome distribution of ChIP-seq peaks identified in PGR-dependent DEG. From (A), peaks that were found in annotated regions belonging to microarray-determined DEG were selected and analysed for genome distribution as defined in Fig. 2. (C) Top most common known motifs found to be enriched at PGR binding sites belonging to PGR-dependent DEG. Bars indicate fold enrichment to background. Circles indicate –log(pvalue). Among motifs found by HOMER that belonged to the same transcription factor family, the most conservative sequences were selected and ranked by fold enrichment compared to background frequency.