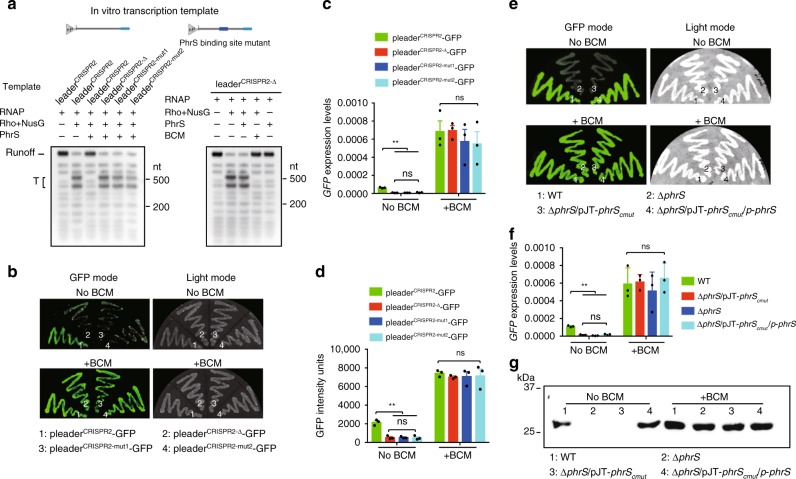
Fig. 6.
The effect of PhrS sRNA on CRISPR transcription via a direct regulatory target. a Transcription reaction shows that PhrS did not hamper Rho termination on CRISPR2 leader mutants’ template with or without BCM. b GFP fluorescence in PA14 strains containing pleaderCRISPR-GFP, pleaderCRISPR-Δ-GFP, pleaderCRISPR-mut1-GFP, or pleaderCRISPR-mut2-GFP plasmid, respectively, without (upper) or with (lower) BCM. c qPCR for GFP levels with or without BCM for PA14 strains containing pleaderCRISPR-GFP, pleaderCRISPR-Δ-GFP, pleaderCRISPR-mut1-GFP, or pleaderCRISPR-mut2-GFP plasmid, respectively. d CRISPR2 leader mutation affect GFP intensity. e GFP fluorescence in PA14 WT, ΔphrS, ΔphrS/pJT-phrScmut, and ΔphrS/pJT-phrScmut/p-phrS strains transformed with pleaderCRISPR-GFP plasmid without (upper) or with (lower) BCM. f qPCR assay for GFP levels without or with BCM for PA14 WT, ΔphrS, ΔphrS/pJT-phrScmut, and ΔphrS/pJT-phrScmut/p-phrS strains transformed with pleaderCRISPR-GFP plasmid. g Western blot analysis of GFP in the PA14 WT, ΔphrS, ΔphrS/pJT-phrScmut, and ΔphrS/pJT-phrScmut/p-phrS strains transformed with pleaderCRISPR-GFP plasmid. Ten micrograms total proteins were used in western blotting analysis. Results are shown with mean ± SEM from three independent experiments. **P < 0.01, *P < 0.05, one-way ANOVA plus Tukey test