Figure 2.
Correcting Mis-estimated CAG Repeat Length Removes Significant Signals at HTT
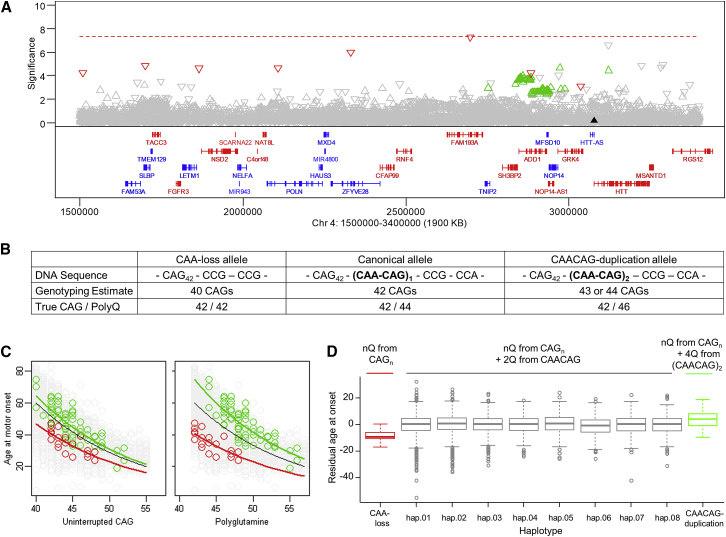
(A) Chr 4 signals (–log10(p value); continuous phenotype) plotted versus genomic coordinate (GRCh37/hg19). The dotted red line indicates genome-wide significance. Red and green symbols are SNPs tagging haplotypes with minor alleles of rs764154313 and rs183415333, respectively. In this and subsequent plots, downward and upward triangles are SNPs with minor alleles associated with hastened and delayed onset, respectively. Genes are below in red (plus strand) and blue (minus strand).
(B) HTT repeat and adjacent sequence, CAG repeat length estimated by genotyping, true CAG repeat length, and polyglutamine length for chrs with CAA-loss (left), canonical (center), and CAACAG-duplication haplotypes, using 42 uninterrupted CAGs as an example.
(C) MiSeq analysis of individuals with rs764154313 and rs183415333 minor alleles identified individuals with CAA-loss (red) and CAACAG-duplication (green) alleles (all others shown in gray), permitting comparison of their age at onset with uninterrupted CAG repeat length (left) or total polyglutamine length (right). The black trend line represents our standard onset-CAG phenotyping model for comparison with trend lines in red and green for those with the CAA-loss and CAACAG-duplication alleles, respectively.
(D) A boxplot (plotted as quartiles: whiskers1.5∗IQR(interquartile range)) of residual age at onset for individuals with a rare CAA-loss HD haplotype (red; N = 21), the 8 most frequent canonical (single CAACAG) HD haplotypes (gray; N = 3357 hap.01, 2016 hap.02, 942 hap.03, 272 hap.04, 266 hap.05, 312 hap.06, 257 hap.07, 302 hap.08) or a rare CAACAG-duplication HD haplotype (green; N = 69), ordered by increasing polyglutamine length (given the same CAG repeat length).