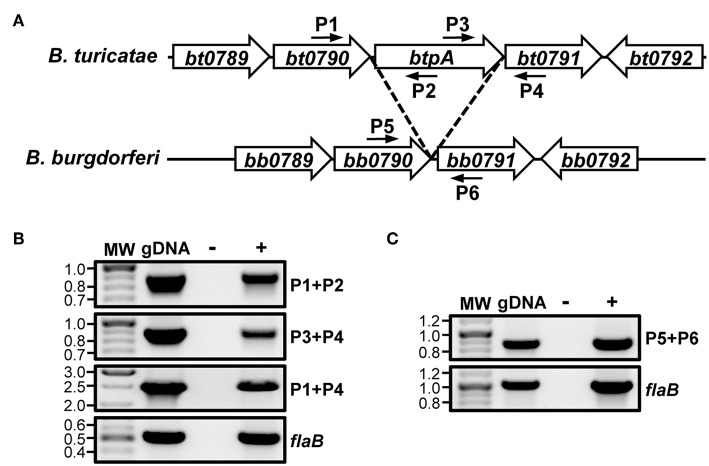
Figure 1.
btpA is transcriptionally linked to bt0790 and bt0791. (A) Selected regions of B. turicatae and B. burgdorferi chromosomes; adapted from Guyard et al. (2006). Numbered smaller arrows indicate primers used in (B,C). (B) Transcriptional linkage of btpA to bt0790 and bt0791 in B. turicatae. RNA was isolated from wild-type B. turicatae culture and converted to cDNA (+). Mock reactions without reverse transcriptase were performed as a negative control (–). Wild-type B. turicatae genomic DNA (gDNA) was included as a positive control for each reaction. Approximate locations of primers in the genome are indicated by smaller arrows and numbers in (A). PCRs were performed to amplify intergenic regions between genes within the suspected operon: bt0790-btpA (P1+P2, 886 bp), btpA-bt0791 (P3+P4, 887 bp), and bt0790-bt0791 (P1+P4, 2,527 bp). Another PCR was performed to amplify an internal region of flaB (519 bp). MW denotes the DNA standard, and numbers to the left indicate molecular weights in kb. (C) Transcriptional linkage of bb0790 and bb0791 in B. burgdorferi. RNA was isolated from B. burgdorferi culture and converted to cDNA (+). Mock reactions without reverse transcriptase were performed as a negative control (–). B. burgdorferi genomic DNA (gDNA) was included as a positive control for each reaction. Approximate locations of primers in the genome are indicated by smaller arrows and numbers in (A). A PCR was performed to amplify an intergenic region between bb0790-bb0791 (P5+P6, 861 bp). Another PCR was performed to amplify an internal region of flaB (1,006 bp). MW denotes the DNA standard, and numbers to the left indicate molecular weights in kb.