Abstract
Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (BMD) are caused by mutations in the DMD gene. The aim of this study is to identify pathogenic DMD variants in probands and reduce the risk of recurrence of the disease in affected families. Variations in 100 unrelated DMD/BMD patients were detected by multiplex ligation-dependent probe amplification (MLPA) and next-generation sequencing (NGS). Pathogenic variants in DMD were successfully identified in all cases, and 11 of them were novel. The most common mutations were intragenic deletions (69%), with two hotspots located in the 5' end (exons 2–19) and the central of the DMD gene (exons 45–55), while point mutations were observed in 22% patients. Further, c.1149+1G>A and c.1150−2A>G were confirmed by hybrid minigene splicing assay (HMSA). This two splice site mutations would lead to two aberrant DMD isoforms which give rise to severely truncated protein. Therefore, the clinical use of MLPA, NGS, and HMSA is an effective strategy to identify variants. Importantly, eight embryos were terminated pregnancies according to prenatal diagnosis and a healthy boy was successfully delivered by preimplantation genetic diagnosis (PGD). Early and accurate genetic diagnosis is essential for prenatal diagnosis/PGD to reduce the risk of recurrence of DMD in affected families.
Keywords: Dystrophin gene, Variation, Genetic diagnosis, Splice site mutation, Hybrid minigene splicing assay
1. Introduction
Dystrophinopathies are common neuromuscular disorders in childhood. There are two main phenotypes: Duchenne muscular dystrophy (DMD, OMIM: 310200) and Becker muscular dystrophy (BMD, OMIM: 300376). DMD, with an approximate incidence of 1/3500 in live male births, is characterized by early-onset, rapidly progressive muscle weakness and being wheel-chair-dependent before age 13, while BMD presents milder clinical features, later-onset and slower progression (Jefferies et al., 2005). Both DMD and BMD are X-linked recessive neuromuscular diseases due to DMD gene (OMIM: 300377) mutations. This is the largest gene in the human genome (Muntoni et al., 2003). The DMD gene spans more than 2.5 million base pairs (bp) of genomic DNA on chromosome X p21.2-21.1 and consists of 79 exons. It encodes a 14 kilobases (kb) mRNA transcript and a 427 kDa full-length dystrophin protein comprising four domains (amino terminal domain, central rod domain, cysteine-rich domain, and carboxy terminal domain) (Muntoni et al., 2003). The dystrophin protein interacts with integral membrane proteins and they assemble into a dystrophin–glycoprotein complex, which plays an important role in stabilizing the sarcolemma and protecting muscle fibers from damage and necrosis (Rando, 2001).
Mutations of DMD include intragenic large deletions/duplications and point mutations. Intragenic deletions and duplications refer to one or more exons (Fairclough et al., 2013), whereas point mutations include nonsense mutations, small frame-shift deletions or insertions, missense, and splice site mutations (Roberts et al., 1994). Multiplex polymerase chain reaction (PCR) is used to be a standard diagnostic method. It could identify 98% of deletions in male patients, but neither duplication and point mutation nor female carriers are detectable (Beggs et al., 1990). Multiplex ligation-dependent probe amplification (MLPA) is currently the most widely used to detect exonic deletions and duplications of DMD both in male patients and female carriers (Schouten et al., 2002; Zimowski et al., 2014). Recently, with the dramatically reduced cost of sequencing, next-generation sequencing (NGS) has become clinically available for the detection of large deletions/duplications and point mutations (nonsense mutation, missense mutation, small insertions/deletions (indels) spanning several base pairs, and splice site mutation) throughout all the exons and flanking introns of DMD (Haas et al., 2011). Splicing regulation has been attracting much more attention since exon-skipping therapy began to appear (Echevarria et al., 2018). So far, a splicing pattern is essential for therapy and diagnosis of disease. Hybrid minigene splicing assay (HMSA) has allowed a relatively fast functional test to build connections between splicing and disease. Though the detection technologies develop rapidly, 1% to 2% of DMD cases remain undiagnosed (Bovolenta et al., 2008).
Identification of pathogenic DMD variants can confirm the diagnosis of DMD/BMD patients and determine the carrier status of their family members. Once the familial mutation is determined, prenatal diagnosis or preimplantation genetic diagnosis (PGD) could be adopted for the family members to prevent the recurrence of the disease. Here, we combined MLPA, NGS, and Sanger sequencing to identify causative mutations in 100 unrelated DMD patients. Eleven novel mutations were detected and two splice site mutations were verified by HMSA. Prenatal diagnosis or PGD was performed on 50 families with known mutations.
2. Materials and methods
2.1. Patients
A total of 100 unrelated DMD/BMD families referred to the Department of Reproductive Genetics, Women’s Hospital, School of Medicine, Zhejiang University (Hangzhou, China) were recruited. All subjects were Han Chinese. The diagnosis of DMD/BMD was based on serum creatine kinase (CK) measurements, muscle biopsy, electromyography, electrocardiogram, progression of the disease, and family history. There were 100 DMD/BMD probands and 100 mothers of probands in our study. This study was approved by the Ethical Committee of the Women’s Hospital, School of Medicine, Zhejiang University. Informed consents were obtained from all participants.
2.2. Genomic DNA extraction
DNA was isolated from peripheral blood lymphocytes and amniocytes according to the manufacturer’s standard protocol (QIAamp DNA Blood Mini Kit, Hilden, Germany). Amniotic fluids were obtained by an obstetrician through amniocentesis and then amniocytes were cultured for eleven days before DNA extraction.
2.3. MLPA
MLPA was used to screen all exons of DMD for all patients. The assay was performed with two independent MLPA kits SALSA P034 and P035 (MRC-Holland, Amsterdam, the Netherlands) according to the manufacturer’s instruction and the PCR products were separated by ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems, CA, USA). Then, the statistical and quantitative analyses were calculated by Coffalyser Net software (http://www.coffalyser.net).
2.4. NGS and Sanger sequencing
NGS was performed as previously described (Wei et al., 2013; Li et al., 2017). All primers were designed by Primer 5.0 (Premier Biosoft International, CA, USA) (Table S1). The sequencing data and the standard DMD reference sequence (GenBank accession number NM_004006.2) were aligned by Mutation Surveyor software (Soft Genetics, State College, PA, USA) to determine nucleotide variations. All mutations identified in this study were checked on the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk www.hgmd.cf.ac.uk), the Leiden Open Variation Database (LOVD, http://www.dmd.nl), and the Universal Mutation Database (UMD, http://www.umd.be). Novel mutations were evaluated according to the American College of Medical Genetics and Genomics (ACMG) guidelines (Richards et al., 2015). Missense mutations were evaluated by Sorts Intolerant from Tolerant (SIFT, http://sift.jcvi.org) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2). Splicing impact was scored using the Human Splicing Finder (HSF, http://www.umd.be/HSF3).
2.5. HMSA
To investigate the effects of the splice site mutations, HMSA including minigene construct, transfection, and reverse transcription PCR (RT-PCR) was performed as previously described (Xia et al., 2018). Human genomic DNA fragments covering from exon 10 to exon 11 were amplified using forward primer DMD-1149/50-F (5'-GATCCTCGAG ctccctattgtctgtatctgc-3') and reverse primer DMD-1149/50-R (5'-GATCGGATCCtacacaatctcctgtgcatgc-3'). The vector primers were as follows: SD6 (5'-TTGTCTACCCATGGACCCAGA-3') and SA2 (5'-CCCCCTGAACCTGAAACATAAAAT-3'). The PCR products were analyzed on 2% (0.02 g/mL) agarose gel and sequencing was performed.
2.6. PGD
In vitro fertilization (IVF) procedure was performed as previously described (Ye et al., 2014). Two or five trophocytes were aspirated from each blastocyst on Day 5 and transferred to PCR tube for whole genome amplification (WGA). One or two non-affected embryos were selected to be transferred. Clinical pregnancy was confirmed by serum β-human chorionic gonadotrophin (β-hCG) and sonography. Amniocentesis was performed at 20 weeks of gestation. Multiple displacement amplification (MDA) was performed according to the manufacturer’s instructions (REPLI-g Single Cell Kit for WGA, QIAGEN, Germany). The MDA products were used for PCR immediately or stored at −20 °C. Preimplantation genetic haplotyping (PGH) was performed by NGS-based single nucleotide polymorphism (SNP) haplotyping. SNP markers located 1 Mb upstream and downstream of the DMD gene in the genomes from the 1000 Genomes Project and known mutation in DMD gene were used for NGS-based SNP haplotyping. The detailed procedure of NGS was performed according to the protocol as previously described (Chen et al., 2016). The data from NGS was further confirmed by standard PCR and Sanger sequencing. Sex determining region Y (SRY) gene was amplified by primers (SRY-F: AGTAAAGGCAACGTCCAGGAT; SRY-R: TTCCGACGAGGTCGATACTTA) and then the amplified products were analyzed by 1.5% (0.015 g/mL) agarose gel and Sanger sequencing.
3. Results
3.1. Deletion and duplication detection
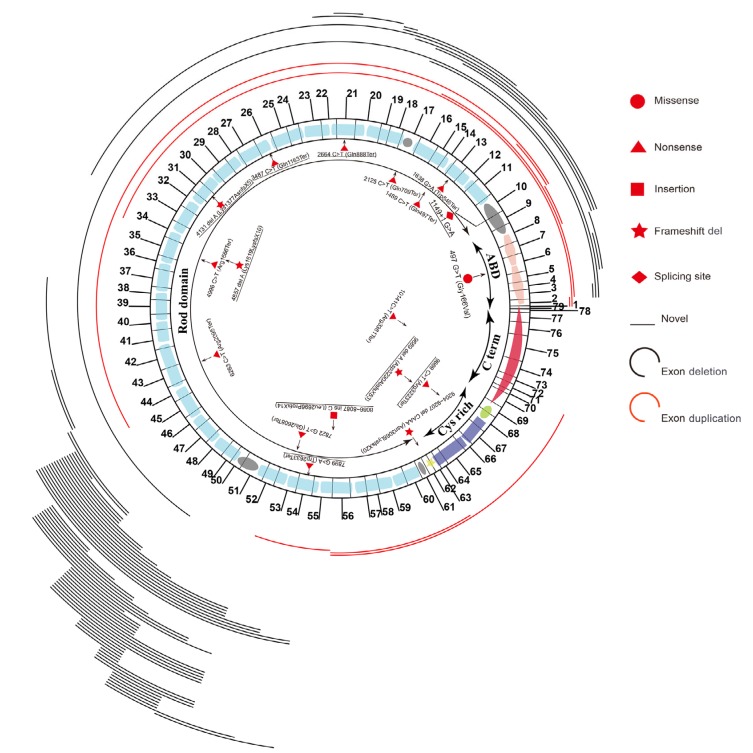
In total, 69 deletions and 9 duplications were identified in this study (Table S2 and Table S3). Notably, two deletion hotspots (exons 45–55 and exons 2–19) were observed (Fig. 1). The number of deleted or duplicated exons ranged widely from 1 to 47. Two or more continuous exon deletions could be easily and reliably determined by MLPA. For the cases with only one exon deletion, the results should be further confirmed by standard PCR and Sanger sequencing. Three-exon deletion/duplication was the most frequent (21.79%, 17/78), followed by single-exon (15.38%, 12/78), five-exon (10.26%, 8/78), and two-exon (8.97%, 7/78). As for the 12 single-exon deletion/duplication mutations, exon 51 was the commonest affected exon (41.67%, 5/12), followed by exon 50 (16.67%, 2/12). Among the 78 affected patients due to deletions/duplications, 47 of them (60.25%, 47/78) inherited the mutation from their mothers, while 31 (39.74%, 31/78) were de novo mutations.
Fig. 1.
Spectrum of DMD mutations identified in this study
Deletions and duplications are located above the schematic representation of exon and dystrophin structural domains of DMD. Black and red lines represent deletion and duplication, respectively. The length of line corresponds to the coverage of deleted and duplicated exons. Point mutations are all located below the schematic representation of exon and dystrophin structural domains of DMD. The types of mutations are distinguished by different symbols (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
3.2. Point mutation detection
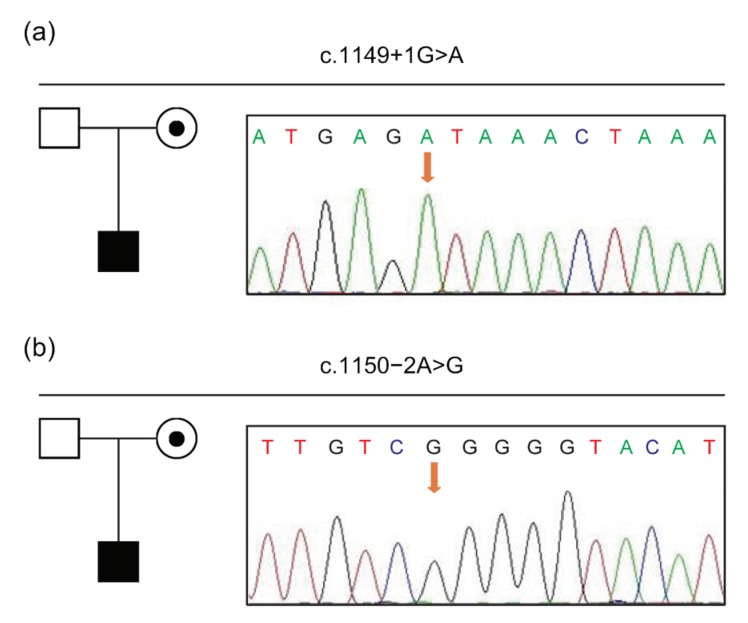
Because of the negative MLPA results of the rest probands, NGS and Sanger sequencing were provided for these subjects. Mutations were found in all 22 probands (Table S4), including 13 nonsense (59.10%, 13/22), 6 indels (27.27%, 6/22), 2 splice site (9.10%, 2/22), and 1 missense (4.55%, 1/22) mutations. Of note, unlike two hotspots observed in deletions, point mutations were scattered throughout the whole DMD gene (Fig. 1). Among them, only three were de novo (13.64%, 3/22). Importantly, two splice site mutations were identified in this study: a novel mutation c.1149+1G>A and a previously reported mutation c.1150−2A>G, which were both inherited from their mothers (Fig. 2). The variation c.1149+1G>A might lead to 5' motif change from GAGgtaaac to GAGataaac, with the wild-type score being 83.90 in comparison with the mutant score of 57.07 as predicted by splicing prediction software HSF. As for c.1150−2A>G, HSF showed the wild-type score was 90.99, while the mutant score was 62.05. Meanwhile, the variation was predicted to break the exonic splicing enhancer (ESE) element and exonic splicing silencer (ESS) element. Therefore, it was suspected that these splice site mutations could break the original splice site. A reported missense mutation c.497G>T was found in a patient. This mutation was predicted to be deleterious by SIFT (affecting protein function with a score of 0.00) and PolyPhen-2 (probably damaging with a score of 1.00).
Fig. 2.
Splice site mutations of DMD gene in the P99 (a) and P100 (b) families
Black boxes indicate affected males; dotted circles indicate female carriers; clear boxes indicate unaffected males. Partial sequencing chromatograms of genomic DNA from patients are shown. The arrows denote the positions of identified mutations
3.3. Splicing study by HMSA
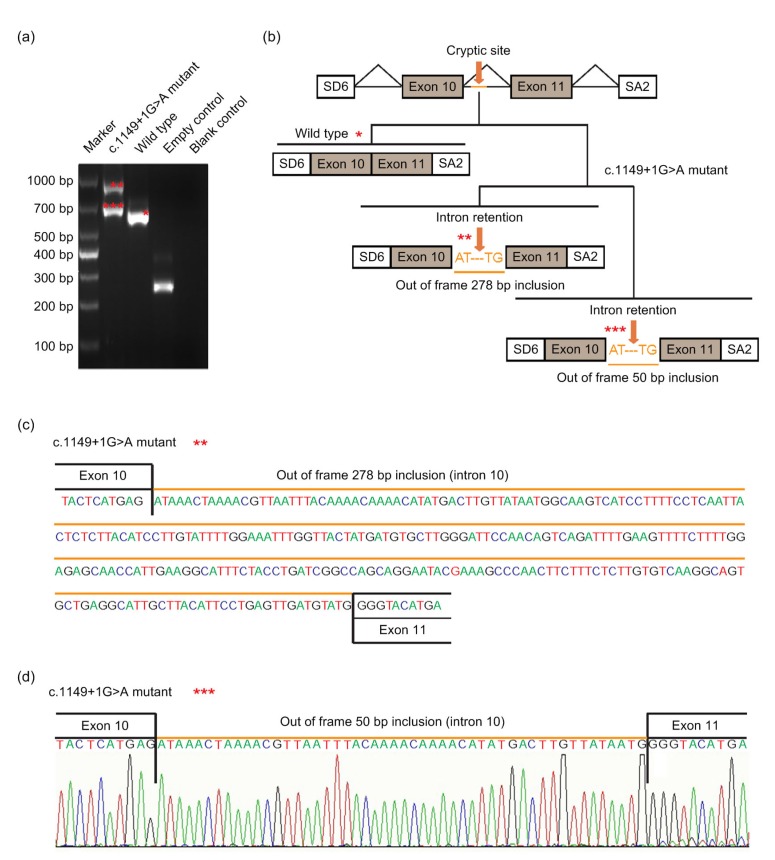
HMSA was performed to investigate the effect of the two splice site mutations. The wide-type minigene construct was 1507 bp, across from partial intron 9 (152 bp), exon 10 (189 bp), intron 10 (650 bp), exon 11 (182 bp) to partial intron 11 (334 bp). A full length of 371 bp RT-PCR product (exclusive SD6 and SA2) was detected, which included exon 10 and exon 11 as expected. There was no product of normal size obtained for the mutant c.1149+1G>A minigene, but instead two abnormal larger fragments of 686 bp and 914 bp were detected resulting from two activations of cryptic donor splice sites in intron 10. Sequencing revealed that the two products contained 50 bp and 278 bp of intron 10, respectively (Fig. 3). They resulted from the activation of a cryptic splice site at different positions in the beginning of intron 10. Theoretically, both mutant isoforms would induce a premature termination codon (PTC) at amino acid 386. The motifs of two cryptic 5' splice sites (5'ss) activated were ATGgcaagt and ATGgtattg, with the score 67.46 and 71.24 calculated by HSF, respectively.
Fig. 3.
Pathogenic analysis of DMD: c.1149+1G>A by HMSA
(a) Electrophoretogram of RT-PCR products. The mutant plasmid showed two types of splicing fragments of 686 bp (***) and 914 bp (**), while the wild-type plasmid showed a normal splicing fragment of 636 bp (*). The empty plasmid including SD6 and SA2 showed 265 bp. (b) Splicing models schematic representation. (c) Sequencing result of the mutated (aberrantly spliced) cDNA showing 278 bp retention of intron 10. (d) Sequencing result of the mutated (aberrantly spliced) cDNA showing 50 bp retention of intron 10
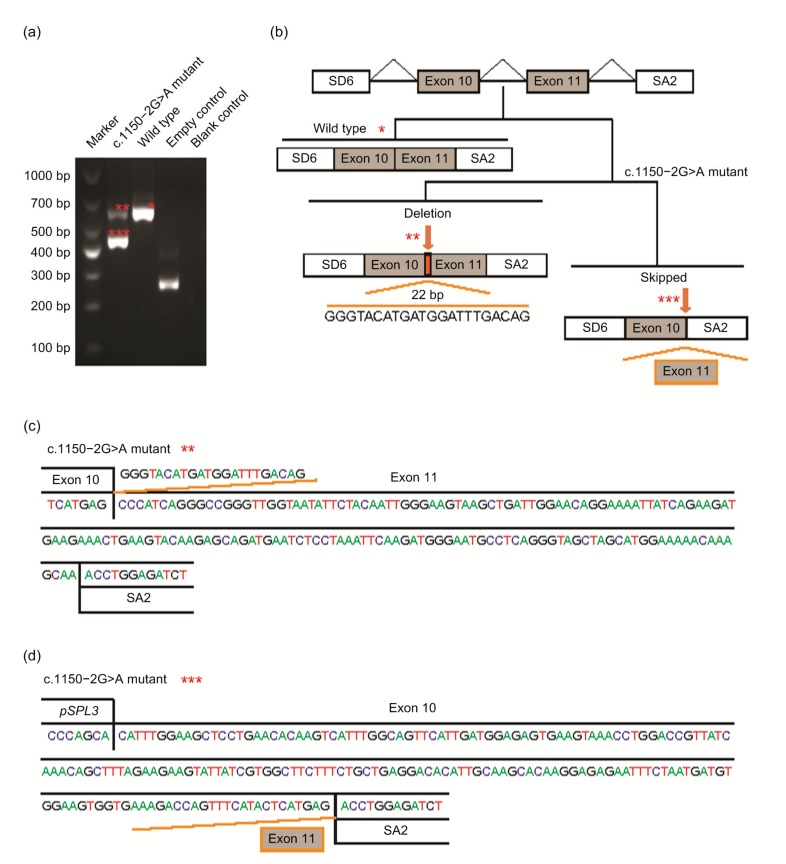
Another minigene construct containing the c.1150−2A>G mutation yielded two shortened transcripts: a primary 454 bp transcript and a 614 bp transcript (Fig. 4). The 454 bp transcript was caused by complete skipping of exon 11 that led to the deletion of 182 bp. Thus, it included exon 10 (189 bp) and 265 bp fragment of exogenous pSPL3 vector by SD6 and SA2 primers. The skipping of exon 11 would cause frameshift that introduced stop codon 9 nucleotides downstream of the junction of exons 10 and 12. The 614 bp transcript resulted from the lack of a part of exon 11 (22 bp), which would cause frameshift that consequently introduced stop codon 46 nucleotides downstream of exon 11. The motif of cryptic 3' splice site (3'ss) activated was ggatttgacagCC, and the score of this cryptic 3'ss was 79.18 calculated by HSF.
Fig. 4.
Pathogenic analysis of DMD: c.1150−2A>G by HMSA
(a) Electrophoretogram of RT-PCR products. The mutant plasmid showed two types of splicing fragments of 454 bp (***) and 614 bp (**), while the wild-type plasmid showed a normal splicing fragment of 636 bp (*). The empty plasmid including SD6 and SA2 showed 265 bp. (b) Splicing models schematic representation. (c) Sequencing result of the mutated (aberrantly spliced) cDNA showing 22 bp lack of exon. (d) Sequencing result of the mutated (aberrantly spliced) cDNA showing skipping of exon 11
3.4. Prenatal diagnosis and PGD
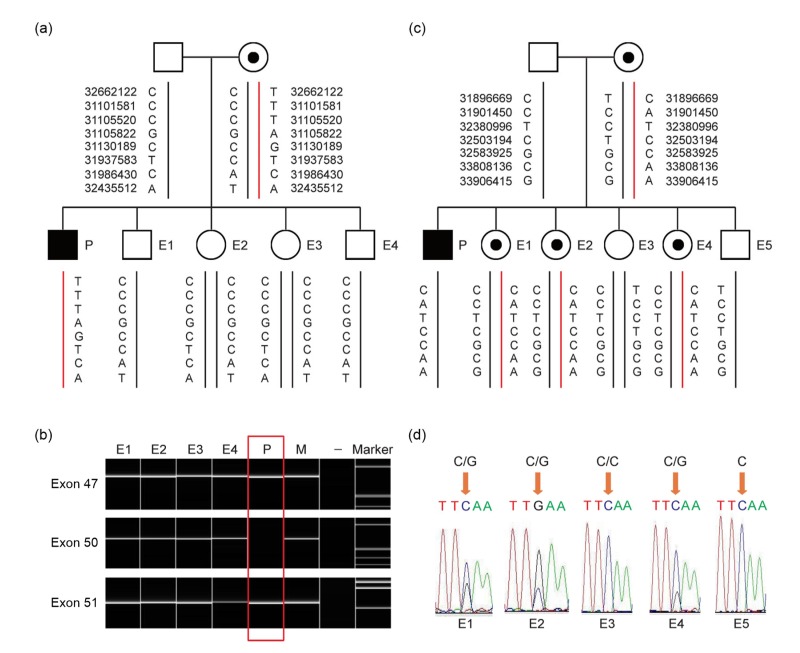
Prenatal diagnosis was performed on 50 mothers of probands (34 carriers and 16 non-carriers). Thirty normal females, 12 normal males, 5 female carriers, and 8 affected males were diagnosed (Table 1). The female carrier of exons 48–50 deletion (P30 family) and female carrier of c.1886C>G (P80 family) underwent PGD by haplotyping and gender determination. For the P30 family, biopsies were undertaken for four embryos: 1 and 4 were male, while embryos 2 and 3 were female. PGH identified 110 SNPs and 8 informative SNPs were selected. PGH and specific detection of deletions showed that all 4 embryos were normal (Fig. 5). Pregnancy was achieved after three cycles of embryo transfer. Amniocyte diagnosis confirmed the PGD results and then a healthy boy was delivered. In the P80 family, embryo biopsies were performed on 5 embryos. PGH identified 91 SNPs and 7 informative SNPs were selected (Fig. 5). Embryos 1, 2, and 4 demonstrated an affected haplotype inherited from the mother, whereas embryos 3 and 5 demonstrated an unaffected haplotype. PGH results were consistent with mutation and sex identification. Results indicated that embryos 1, 2, and 4 were female carriers, while embryos 3 and 5 were normal female and male, respectively. Finally, one embryo was transferred, resulting in a biochemical pregnancy. At 20 weeks of gestation, amniocyte diagnosis was performed and confirmed the PGD results.
Table 1.
Prenatal diagnosis performed on 50 mothers of probands
| Family No. | Carrier status of mother | Prenatal diagnosis |
| P4 | Yes | NF |
| P91 | No | dNF |
| P27 | No | NF |
| P35 | No | NF |
| P36 | No | NF |
| P48 | No | NM |
| P51 | No | NF |
| P52 | No | NF |
| P74 | No | NM |
| P56 | No | NF |
| P57 | No | NF |
| P60 | No | NF |
| P62 | Yes | NF |
| P63 | Yes | NM |
| P65 | Yes | Carrier |
| P67 | Yes | Carrier |
| P68 | Yes | NF |
| P11 | Yes | NM |
| P7 | No | NF |
| P40 | No | NF |
| P84 | Yes | Patient |
| P46 | No | NF1/NM2 |
| P69 | No | NF |
| P16 | Yes | Patient |
| P20 | Yes | NF |
| P86 | Yes | Patient |
| P92 | Yes | Patient |
| P94 | Yes | NF |
| P95 | Yes | NM |
| P96 | Yes | NF |
| P97 | Yes | Patient1/Carrier2 |
| P31 | Yes | NM |
| P39 | Yes | dNF |
| P73 | Yes | NF |
| P43 | Yes | Patient |
| P49 | Yes | Carrier |
| P50 | Yes | Patient |
| P58 | Yes | NF |
| P77 | Yes | NM |
| P30 | Yes | NM |
| P80 | Yes | NF |
| P18 | Yes | NF |
| P34 | Yes | NF |
| P8 | Yes | NM |
| P15 | No | NM |
| P33 | Yes | Patient1/NM2 |
| P12 | Yes | NF |
| P32 | Yes | NF |
| P76 | Yes | NF |
| P82 | Yes | Carrier |
1/2: number of pregnance; NF: normal female fetuses; NM: normal male fetuses; d: double fetuses
Fig. 5.
PGD results of P30 and P80 families
(a) Haplotype analysis of P30 family. Diagram shows that all embryos have normal haplotype. (b) Direct mutation detection by amplification of exons 47, 50, and 51 of DMD for P30 family. All embryos had no deletion of exon 50. (c) Haplotype analysis of P80 family. E1, E2, and E4 demonstrated an affected haplotype pattern (red-line allele) inherited from mother, whereas E3 and E5 demonstrated an unaffected haplotype pattern (black-line allele). (d) Direct detection c.1886C/C in exon 16 of DMD for P80 family. E3 had wild homozygote c.1886C/C, whereas E5 had hemizygote. However, E1, E2, and E4 had heterozygous c.1886C>G (p.Ser629X). P: proband; M: proband’s mother; E1−E5: embryos 1–5; −: negative control. Black boxes indicate affected males; dotted circles indicate female carriers; clear boxes and circles indicate unaffected males and females, respectively (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
4. Discussion
In this study, all pathogenic variants in DMD were successfully identified in all 100 cases, and 11 variants were novel (Tables 2 and 3). Nonsense, indels and splice site mutations at ±1/±2 were pathogenic, as evaluated according to ACMG guidelines (Richards, et al., 2015). Our findings expanded the spectrum of the Human Mutation Database. MLPA was used as the first-line screening method and our results showed that the percentage of deletion and duplication detection by MLPA is 78%, which was similar to other studies (70%) (Lalic et al., 2005; Takeshima et al., 2010). As for the cases with negative MLPA results, we further used NGS to detect possible point mutations (Flanigan et al., 2003). In total, 22 point mutations out of 100 cases (22%) were identified. Point mutations were dominated by nonsense mutation, which was consistent with previous studies (Tuffery-Giraud et al., 2009; Takeshima et al., 2010).
Table 2.
Characteristics of the detected mutations
| Mutation type | Pathogenic | Known | Novel | De novo | Hereditary |
| Deletion | 69 | 67 | 2 | 29 | 40 |
| Duplication | 9 | 8 | 1 | 2 | 7 |
| Nonsense | 13 | 9 | 4 | 3 | 10 |
| Splicing | 2 | 1 | 1 | 0 | 2 |
| Missense | 1 | 1 | 0 | 0 | 1 |
| Indels | 6 | 3 | 3 | 0 | 6 |
|
| |||||
| Total | 100 | 89 | 11 | 34 | 66 |
Table 3.
Eleven novel variants detected in this study
| Patient No. | Variant | Inheritance |
| P4 | EX5–6 deletion | Maternal |
| P43 | EX12–33 deletion | Maternal |
| P77 | EX56–70 duplication | Maternal |
| P79 | c.3487C>T, p.(Gln1163Ter) | De novo |
| P82 | c.8086_8087insC, p.(Leu2696ProfsX14) | Maternal |
| P85 | c.9689delA, p.(Asp3230AlafsX53) | Maternal |
| P88 | c.9669C>T, p.(Arg3223Ter) | Maternal |
| P89 | c.2664 C>T, p.(Gln888Ter) | De novo |
| P92 | c.7899G>A, p.(Trp2633Ter) | Maternal |
| P94 | c.4131delA, p.(Lys1377AsnfsX5) | Maternal |
| P99 | c.1149+1G>A | Maternal |
We identified a potential pathogenic missense mutation c.497G>T (p.Gly166Val) which was reported previously (Juan-Mateu et al., 2013). This mutation was located at highly conserved residues and was predicted to be pathogenic. In pedigree 81, there were three affected males who had hemizygote c.497G>T. The first affected male had severe muscle weakness and died from respiratory failure at 20 years of age. The second one was early-onset and lost ambulation as disease progressed at 15 years of age, while the third one was 7-year-old now and suffered from progressive muscle weakness from 4 years of age. Juan-Mateu et al. (2013) had reported that mutation c.497G>T was found in an intermediate muscular dystrophy patient (loss of ambulation between 13 and 16 years of age), which was almost consistent with the patient’s clinical phenotype in this study.
In DMD patients, prediction of splicing outcomes is essential for determining the phenotype. As for novel mutation c.1149+1G>A, the HMSA results revealed that two abnormal transcripts of 50 bp and 278 bp retention partial intron 10 resulted from the activation of cryptic donor splice sites in intron 10 and both of them were out of frame. This was consistent with the patient’s DMD phenotype (early-onset and wheel-chair-dependent at 8 years of age). The mutation c.1150−2A>G was previously reported once in a DMD case (Ma et al., 2018). However, no experimental evidence was available for this mutation. In our study, we not only verified that the mutation was maternal but the splicing outcome that arose from the mutation was conducted by HMSA. It revealed that two abnormal transcripts of complete skipping of exon 11 and lack of 22 bp of exon 11. These two abnormal splicing outcomes were out of frame, which was consistent with the patient’s DMD phenotype (early-onset and wheel-chair-dependent at 9 years of age).
Mutations involving GT and AG dinucleotide at the 5' (donor) and 3' (acceptor) splice sites would lead to disease (Pico et al., 2014). The major consequences of mutation at splice sites were exon skipping and cryptic splice site activation (Baralle and Baralle, 2005). The decision between two patterns of splicing outcomes was rather a matter of exon and intron length, exon flanking sequence, RNA secondary structure, open reading frame conservation, and DNA sequence environment (Roca et al., 2003; Buratti and Baralle, 2004; Zhang et al., 2005; Buratti et al., 2006; Hertel, 2008; Pico et al., 2014). Exon skipping was more common than cryptic donor splice site activation in the dystrophin gene mutations in cases of intron +1G>A, while cryptic splice site activation was likely only seen in strong exons (>170 bp) (Habara et al., 2009). As for the c.1149+1G>A case, it was suspected that cryptic splice site activation was influenced by the long length of exon 10 (189 bp). Minigene assay identified two cryptic 5'ss, and the scores calculated by HSF were lower than that of wild-type 5'ss, but higher than that of the mutant one. Actually, several potential 5'ss predicted by HSF were between the mutated authentic splice site and cryptic 5'ss. These potential 5'ss were completely silent and likely to be pseudo 5'ss. Roca et al. (2003) observed that the cryptic 5'ss activated were usually located close to the authentic 5'ss, whereas a few of cryptic 5'ss located further away were also activated. We suggested that the distance to the mutated splice site played an important role, but not the decisive factor for cryptic 5'ss activation. However, we observed the difference in splicing efficiency between these two cryptic 5'ss, which might be associated with proximity to the mutated splice site. It was interesting to note that one of cryptic splice sites activated at 50 bp downstream of exon 10 was activated, though the following nucleotides are “GC.” The GT/AG splice sites were present in the majority of eukaryotic introns. In a database (SpliceDB, http://www.softberry.com/spldb/SpliceDB.html) of known mammalian splice sites, Burset et al. (2001) observed that non-canonical GC sequence at 5'ss accounted for 0.56% and GC 5'ss possess a strong consensus sequence. Parada et al. (2014) uncovered that the GC/AG splice sites comprise about 0.889% of the total splice sites in humans. Although the percentage of non-canonical GC 5'ss was low, the GC 5'ss existed in various species, and were not aberrant splice sites. Also, a GC/AG splice sites pair existed at intron 30 of DMD. The factors promoting such weak site efficient selection are still poorly understood. Kralovicova et al. (2011) showed high density of GAA/CAA with splicing enhancers in exon making a contribution for efficient selection of GC 5'ss. In the absence of an alternative splice site, exon skipping was the preferred outcome at both donor and acceptor splice sites (Krawczak et al., 2007). As for c.1150−2A>G, exon 11 skipping was the major splicing outcome, and in contrast to the cryptic splice site was activated slightly. The sub-optimal, neighboring 3'ss activated was tggatttgacag/CC and the score calculated by HSF was lower for this cryptic 3'ss with respect to wild-type 3'ss. The variation c.1150−2A>G was predicted to break ESE and ESS, which regulate premature mRNA splicing. These regulatory elements may play important roles in the pre-mRNA splicing process and have an impact on transcriptional efficiency. It was supposed that skipping of the adjacent exon was the preferred outcome when cryptic splice site was of low efficiency or not sufficient. So far, it is still a challenge to establish a rule to predict splicing outcomes. Though HMSA is an effective validation technique in the molecular laboratory, the appropriate RNA analysis ideally comes from affected tissue because of the tissue-specific dystrophin functions (Muntoni et al., 2003). So it is important to keep in mind that the splicing outcome would be different in artificial minigenes as compared to endogenous transcripts.
Early diagnosis of DMD and genetic confirmation of the causative mutation in patients are important. Once a mutation is identified, carrier analysis of female relatives can be performed. In our study, DMD gene mutation was inherited from the mother in 66% (66/100) of cases, whereas the remaining 34% (34/100) were de novo, which was consistent with a previous report (30%) (Sakthivel Murugan et al., 2013). However, it should be noted that a mother without any mutation in her peripheral blood lymphocytes is still at risk due to the possibility of germinal mosaicism (Bakker et al., 1989; van Essen et al., 1992).
Prenatal diagnosis and PGD are valid tools to reduce the risk of having affected children. In 50 families with confirmed mutations, prenatal diagnoses were successfully performed by MLPA or Sanger sequencing. Eight fetuses were verified as affected boy, approximately theoretical value 25%, and five fetuses were carriers in 34 mother carriers. On the other hand, all fetuses were identified as unaffected in 16 mothers without mutation in their peripheral blood lymphocytes. In P30 family with deletion of exons 48–50, PGD was performed and a healthy boy was successfully delivered. Therefore, it is obvious that PGD is an alternative to prenatal diagnosis for a family at high risk of transmitting an inherited disease to offspring and avoiding the possible termination of pregnancy. Specific mutation detection (Girardet et al., 2003) and haplotype analysis (Lee et al., 1998) with gender determination have been universally used for PGD of DMD.
In conclusion, it is a great challenge to perform comprehensive analyses of causative mutations in DMD because of its large size and the variety of mutation types (Stockley et al., 2006). Our study provided a comprehensive combination of MLPA, NGS, and HMSA, which is an effective strategy to identify DMD pathogenic variants. Early and accurate genetic diagnosis and confirmation of variants are essential for therapy and prenatal diagnosis/PGD to reduce the risk of recurrence of DMD in affected families.
Acknowledgments
The authors thank the patients for their voluntary involvement in this study. We are grateful to Dr. Jiong GAO (BGI Genomics, Shenzhen, China) for analyzing the data and modifying the article.
List of electronic supplementary materials
Oligonucleotide primers for detection of mutations in DMD gene
Deletions in DMD gene detected by MLPA
Duplications in DMD gene detected by MLPA
Point mutations in DMD gene detected by NGS
Footnotes
Project supported by the National Key Research and Development Program of China (No. 2016YFC1000703), the Medicine and Health Science and Technology Plan Projects in Zhejiang Province (No. 2014KYA246), and the National Natural Science Foundation of China (Nos. 81801441 and 81300532)
Contributors: Min-yue DONG set up the theme and examined the manuscript. Yan-mei YANG performed the experiments and wrote the manuscript. Kai YAN, Bei LIU, Li-ya WANG, and Ying-zhi HUANG performed the experiments. Min CHEN was responsible for editing the grammar of the manuscript. Ye-qing QIAN guided the modification process. Yi-xi SUN and Hong-ge LI were responsible for collecting the patients. All authors read and approved the final manuscript and, therefore, had full access to all the data in the study and take responsibility for the integrity and security of the data.
Electronic supplementary materials: The online version of this article (https://doi.org/10.1631/jzus.B1800541) contains supplementary materials, which are available to authorized users
Compliance with ethics guidelines: Yan-mei YANG, Kai YAN, Bei LIU, Min CHEN, Li-ya WANG, Ying-zhi HUANG, Ye-qing QIAN, Yi-xi SUN, Hong-ge LI, and Min-yue DONG declare that they have no conflict of interest.
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study. Additional informed consent was obtained from all patients for whom identifying information is included in this article.
References
- 1.Bakker E, Veenema H, den Dunnen JT, et al. Germinal mosaicism increases the recurrence risk for ‘new’ Duchenne muscular dystrophy mutations. J Med Genet. 1989;26(9):553–559. doi: 10.1136/jmg.26.9.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42(10):737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beggs AH, Koenig M, Boyce FM, et al. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet. 1990;86(1):45–48. doi: 10.1007/BF00205170. [DOI] [PubMed] [Google Scholar]
- 4.Bovolenta M, Neri M, Fini S, et al. A novel custom high density-comparative genomic hybridization array detects common rearrangements as well as deep intronic mutations in dystrophinopathies. BMC Genomics, 9:572. 2008 doi: 10.1186/1471-2164-9-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24(24):10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34(12):3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burset M, Seledtsov IA, Solovyev VV. SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 2001;29(1):255–259. doi: 10.1093/nar/29.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LJ, Diao ZY, Xu ZP, et al. The clinical application of preimplantation genetic diagnosis for the patient affected by congenital contractural arachnodactyly and spinal and bulbar muscular atrophy. J Assist Reprod Genet. 2016;33(11):1459–1466. doi: 10.1007/s10815-016-0760-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echevarria L, Aupy P, Goyenvalle A. Exon-skipping advances for Duchenne muscular dystrophy. Hum Mol Genet. 2018;27(R2):R163–R172. doi: 10.1093/hmg/ddy171. [DOI] [PubMed] [Google Scholar]
- 10.Fairclough RJ, Wood MJ, Davies KE. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat Rev Genet. 2013;14(6):373–378. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 11.Flanigan KM, von Niederhausern A, Dunn DM, et al. Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet. 2003;72(4):931–939. doi: 10.1086/374176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girardet A, Hamamah S, Dechaud H, et al. Specific detection of deleted and non-deleted dystrophin exons together with gender assignment in preimplantation genetic diagnosis of Duchenne muscular dystrophy. Mol Hum Reprod. 2003;9(7):421–427. doi: 10.1093/molehr/gag050. [DOI] [PubMed] [Google Scholar]
- 13.Haas J, Katus HA, Meder B. Next-generation sequencing entering the clinical arena. Mol Cell Probes. 2011;25(5-6):206–211. doi: 10.1016/j.mcp.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Habara Y, Takeshima Y, Awano H, et al. In vitro splicing analysis showed that availability of a cryptic splice site is not a determinant for alternative splicing patterns caused by +1G→A mutations in introns of the dystrophin gene. J Med Genet. 2009;46(8):542–547. doi: 10.1136/jmg.2008.061259. [DOI] [PubMed] [Google Scholar]
- 15.Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283(3):1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- 16.Jefferies JL, Eidem BW, Belmont JW, et al. Genetic predictors and remodeling of dilated cardiomyopathy in muscular dystrophy. Circulation. 2005;112(18):2799–2804. doi: 10.1161/CIRCULATIONAHA.104.528281. [DOI] [PubMed] [Google Scholar]
- 17.Juan-Mateu J, González-Quereda L, Rodríguez MJ, et al. Interplay between DMD point mutations and splicing signals in dystrophinopathy phenotypes. PLoS ONE. 2013;8(3):e59916. doi: 10.1371/journal.pone.0059916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kralovicova J, Hwang G, Asplund AC, et al. Compensatory signals associated with the activation of human GC 5' splice sites. Nucleic Acids Res. 2011;39(16):7077–7091. doi: 10.1093/nar/gkr306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krawczak M, Thomas NST, Hundrieser B, et al. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat. 2007;28(2):150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 20.Lalic T, Vossen RHAM, Coffa J, et al. Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet. 2005;13(11):1231–1234. doi: 10.1038/sj.ejhg.5201465. [DOI] [PubMed] [Google Scholar]
- 21.Lee SH, Kwak IP, Cha KE, et al. Preimplantation diagnosis of non-deletion Duchenne muscular dystrophy (DMD) by linkage polymerase chain reaction analysis. Mol Hum Reprod. 1998;4(4):345–349. doi: 10.1093/molehr/4.4.345. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Pan Q, Gu YS. Phenotype-genotype correlation with Sanger sequencing identified retinol dehydrogenase 12 (RDH12) compound heterozygous variants in a Chinese family with Leber congenital amaurosis. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2017;18(5):421–429. doi: 10.1631/jzus.B1600156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma PP, Zhang S, Zhang H, et al. Comprehensive genetic characteristics of dystrophinopathies in China. Orphanet J Rare Dis, 13:109. 2018 doi: 10.1186/s13023-018-0853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2(12):731–740. doi: 10.1016/S1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 25.Parada GE, Munita R, Cerda CA, et al. A comprehensive survey of non-canonical splice sites in the human transcriptome. Nucleic Acids Res. 2014;42(16):10564–10578. doi: 10.1093/nar/gku744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pico AR, Bader GD, Demchak B, et al. The Cytoscape app article collection. F1000Res, 3:138. 2014 doi: 10.12688/f1000research.4642.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rando TA. The dystrophin–glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve. 2001;24(12):1575–1594. doi: 10.1002/mus.1192. [DOI] [PubMed] [Google Scholar]
- 28.Roberts RG, Gardner RJ, Bobrow M. Searching for the 1 in 2,400,000: a review of dystrophin gene point mutations. Hum Mutat. 1994;4(1):1–11. doi: 10.1002/humu.1380040102. [DOI] [PubMed] [Google Scholar]
- 29.Roca X, Sachidanandam R, Krainer AR. Intrinsic differences between authentic and cryptic 5' splice sites. Nucleic Acids Res. 2003;31(21):6321–6333. doi: 10.1093/nar/gkg830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakthivel Murugan SM, Arthi C, Thilothammal N, et al. Carrier detection in Duchenne muscular dystrophy using molecular methods. Indian J Med Res. 2013;137(6):1102–1110. [PMC free article] [PubMed] [Google Scholar]
- 32.Schouten JP, McElgunn CJ, Waaijer R, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stockley TL, Akber S, Bulgin N, et al. Strategy for comprehensive molecular testing for Duchenne and Becker muscular dystrophies. Genet Test. 2006;10(4):229–243. doi: 10.1089/gte.2006.10.229. [DOI] [PubMed] [Google Scholar]
- 34.Takeshima Y, Yagi M, Okizuka Y, et al. Mutation spectrum of the dystrophin gene in 442 Duchenne/Becker muscular dystrophy cases from one Japanese referral center. J Hum Genet. 2010;55(6):379–388. doi: 10.1038/jhg.2010.49. [DOI] [PubMed] [Google Scholar]
- 35.Tuffery-Giraud S, Béroud C, Leturcq F, et al. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD–DMD database: a model of nationwide knowledgebase. Hum Mutat. 2009;30(6):934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 36.van Essen AJ, Abbs S, Baiget M, et al. Parental origin and germline mosaicism of deletions and duplications of the dystrophin gene: a European study. Hum Genet. 1992;88(3):249–257. doi: 10.1007/BF00197255. [DOI] [PubMed] [Google Scholar]
- 37.Wei XM, Dai Y, Yu P, et al. Targeted next-generation sequencing as a comprehensive test for patients with and female carriers of DMD/BMD: a multi-population diagnostic study. Eur J Hum Genet. 2013;22(1):110–118. doi: 10.1038/ejhg.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia ZJ, Lin J, Lu LP, et al. An intronic mutation c.6430-3C>G in the F8 gene causes splicing efficiency and premature termination in hemophilia A. Blood Coagul Fibrinoly. 2018;29(4):381–386. doi: 10.1097/MBC.0000000000000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye Y, Yu P, Yong J, et al. Preimplantational genetic diagnosis and mutation detection in a family with duplication mutation of DMD gene. Gynecol Obstet Invest. 2014;78(4):272–278. doi: 10.1159/000365083. [DOI] [PubMed] [Google Scholar]
- 40.Zhang XHF, Leslie CS, Chasin LA. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15(6):768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimowski JG, Massalska D, Holding M, et al. MLPA based detection of mutations in the dystrophin gene of 180 Polish families with Duchenne/Becker muscular dystrophy. Neurol Neurochir Pol. 2014;48(6):416–422. doi: 10.1016/j.pjnns.2014.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers for detection of mutations in DMD gene
Deletions in DMD gene detected by MLPA
Duplications in DMD gene detected by MLPA
Point mutations in DMD gene detected by NGS