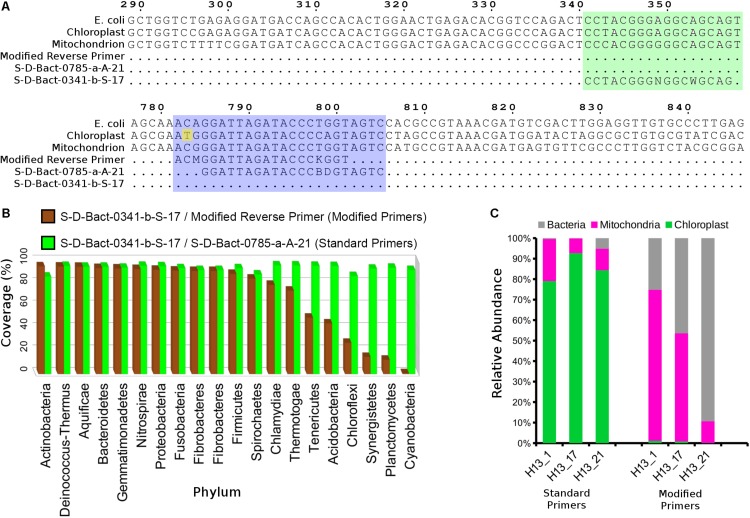
FIGURE 1.
Modification of the 16S V3–V4 reverse primer reduced co-amplification of grapevine rRNA gene. (A) Nucleotide alignment of standard and modified primers against the grapevine mitochondrial and chloroplast rRNA genes. Green and purple blocks indicate 16S rRNA gene regions containing the aligned forward and reverse 16S V3–V4 primers, respectively. Numbers above the alignment indicate base position on the Escherichia coli 16S rRNA gene sequence. (B) Phylum-level taxonomic coverage of the standard and modified V3–V4 primer pairs as assessed by SILVA TestPrimer 1.0 based on the SILVA SSU r132 RefNR database (maximum number of mismatches = 5; length of 0-mismatch zone at 3′-end = 5 bases). (C) Relative abundance of bacterial, grapevine mitochondrial, and chloroplast sequence for three 2013 Hungarian crown gall samples that were amplified using the standard and modified 16S V3–V4 primers.