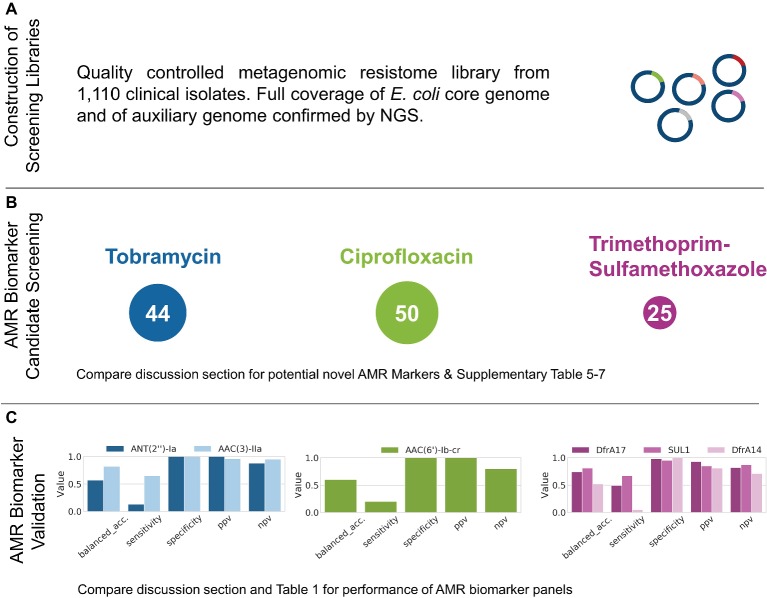
Figure 2.
Results obtained from the functional metagenomics screening workflow described in Figure 1. After Step 5, the representative coverage of the 1,110 E. coli genomes by the metagenomic library was confirmed by NGS (A). AMR screening after Step 8 identified 44, 50, and 25 AMR marker candidates for TOB, CIP, and TMP-SMX, respectively (B). After further prioritization based on matched genotype-phenotype data for the isolates used for library generation, the proposed workflow was applied to successfully recover and validate AMR markers for all antibiotics tested (Step 10, C).