Abstract
Cancer cells generally harbor hundreds of alterations in the cancer genomes and act as crucial factors in the development and progression of cancer. Gene alterations in the cancer genome form genetic interactions, which affect the response of patients to drugs. We developed an algorithm that mines copy number alteration and whole-exome mutation profiles from The Cancer Genome Atlas (TCGA), as well as functional screen data generated to identify potential genetic interactions for specific cancer types. As a result, 4,529 synthetic viability (SV) interactions and 10,637 synthetic lethality (SL) interactions were detected. The pharmacogenomic datasets revealed that SV interactions induced drug resistance in cancer cells and that SL interactions mediated drug sensitivity in cancer cells. Deletions of HDAC1 and DVL1, both of which participate in the Notch signaling pathway, had an SV effect in cancer cells, and deletion of DVL1 induced resistance to HDAC1 inhibitors in cancer cells. In addition, patients with low expression of both HDAC1 and DVL1 had poor prognosis. Finally, by integrating current reported genetic interactions from other studies, the Cancer Genetic Interaction database (CGIdb) (http://www.medsysbio.org/CGIdb) was constructed, providing a convenient retrieval for genetic interactions in cancer.
Keywords: synthetic viability, synthetic lethality, drug resistance, drug sensitivity, biomarker
Introduction
Traditional cancer therapy predominantly involves chemotherapy, which provides limited improvements in patient survival. Patients frequently acquire resistance to chemotherapy, significantly decreasing the usefulness of anticancer drugs in the clinic.1 As a new approach of precision medicine, target therapy has brought a new opportunity for cancer treatment. Target therapy refers to formulating the best treatment regimens for patients according to personalized genome characteristics, maximizing the treatment effect and minimizing patient injury.2 There are hundreds of genomic alterations in tumor cells, such as copy number alterations (CNAs) and somatic mutations. Cancer patients, even with the same cancer type, have high heterogeneous genomic alterations,3 which induce complex drug responses. It is still a great challenge to identify effective biomarkers to predict drug resistance and drug sensitivity for target therapy. Inevitable drug resistance in target therapy is also a major obstacle. Tremendous efforts and progress have been made over the past few years to reveal the specific mechanism of drug resistance and sensitivity, including genetic interactions among gene alterations in the cancer genome.4, 5
The generation of an unexpected phenotypic outcome when combining two alterations is referred to as a genetic interaction, including synthetic viability and synthetic lethality.6 Synthetic viability (SV) describes the scenario in which the synthesis or combination of two gene effects remedy the effect of single-gene defects, such as cell death or a significant impairment of fitness, and enhance cell survival, which may be a potential mechanism for drug resistance.5 Synthetic lethality (SL) describes the scenario in which single-gene defects are compatible with cell viability, but the combination of alterations in both genes induces cell death, which is generally used to identify drug-sensitive biomarkers or drug targets.7 A well-known example of an SL effect is the selective impediment of PARP inhibitors on the growth of BRCA1/2-mutated cancer cells, and this SL effect has been successfully applied to clinical therapy.8 Olaparib was approved by the US Food and Drug Administration (FDA) for treatment of BRCA1/2-mutated advanced ovarian cancer patients in 2014. However, many ovarian cancer patients carrying mutations of BRCA1/2 have resistance to olaparib in clinical applications.9 Hu et al.10 found that GPBP1 loss causes resistance to PARP inhibitors by regulating the expression of factors involved in homologous recombination (HR) according to a quantitative chemotherapy genetic interaction map.
Gene alterations in the cancer genome present specific patterns, such as co-occurrence or mutual exclusivity.11 Mutual exclusivity indicates that both gene alterations rarely occur in the same patients, and mutual exclusivity has been used to identify synthetic lethal interactions in cancer cells. For example, Unni et al.12 reported that an SL effect underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. Co-occurring alterations, in which both gene alterations significantly and simultaneously occur in cancer patients, may have an SV effect on cancer cells, and these alterations have been validated in our previous work.5 Recent advances in functional screening technology provide an avenue to explore genetic interactions in cancer cells.13 However, most studies have focused on the identification of SL interactions and further detection of drug targets while ignoring SV interactions involved in the cancer genome. Thus, besides SL interactions, the present work identified and analyzed SV interactions in cancer cells using The Cancer Genome Atlas (TCGA) cancer genome data and validating by short hairpin RNA (shRNA), CRISPR, and yeast datasets. Moreover, the present study further detected drug-resistance biomarkers using SV interactions based on the hypothesis that cancer cells with alterations in partner genes are resistant to specific drugs if drug targets and partner genes have SV effects.
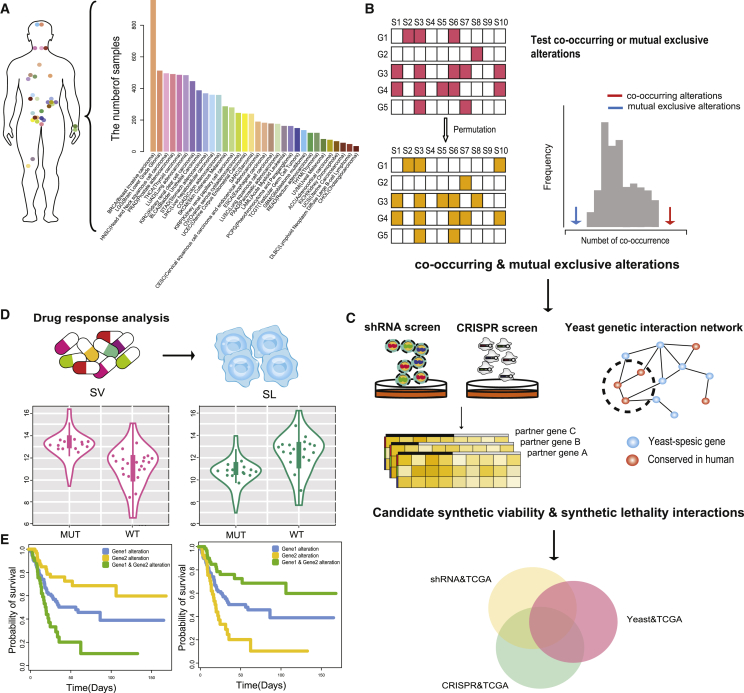
In the present study, CNAs and the mutation and expression profiles of 8,580 samples were integrated across 32 cancer types from TCGA to identify co-occurrence and mutual exclusivity gene pairs (Figures 1A and 1B). Reliable co-occurrence and mutual exclusivity interactions were then selected from functional screening data, including shRNA and CRISPR datasets (Figure 1C). The strategy was based on the notion that knockdown of one gene causes a selective enhancement (SV) or reduction (SL) in cell viability, with simultaneous alterations in another gene. In addition, the positive and negative interactions in yeast genetic interactions were also used to infer SV and SL relationships in human cancer cells, respectively. Ultimately, the co-occurrence and mutual exclusivity gene pairs verified in at least one type of dataset (shRNA, CRISPR, or yeast) were selected as candidate genetic interactions (Figure 1C). SV and SL interactions were validated by (1) showing the expected drug response detected by four pharmacogenomic datasets (Figure 1D) as well as (2) observing worse survival of patients with alterations in SV interactions and better survival of patients with alterations in SL interactions in the prognosis analysis (Figure 1E). Moreover, network analysis and pathway enrichment analysis were performed to investigate the functional relationship between genes with genetic interactions. The biomarkers identified by the present work will contribute to predict the mechanism of drug resistance or sensitivity in clinical application and will guide precise targeting of existing therapies.
Figure 1.
Workflow of the Present Study
(A) Statistics of 32 cancers in TCGA. (B) Identification of co-occurring and mutual exclusive interactions across 32 cancer types. (C) Identification of candidate genetic interactions using shRNA, CRISPR screening data, and yeast genetic interaction data. (D) Prediction of biomarkers for cancer cell drug sensitivity and resistance. (E) Prognostic analysis.
Results
Identification of Candidate SV Interactions and SL Interactions in Cancers
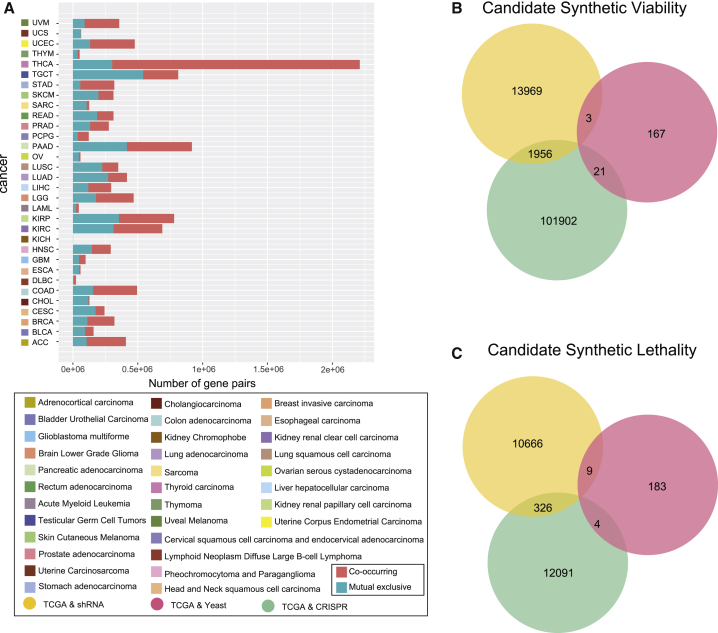
To identify significant co-occurring and mutual exclusive relationships among genomic alterations, mutation and CNA data from 8,580 samples of 32 different cancer types studied by the TCGA consortium were utilized (Figure 1A). Considering the heterogeneous distribution of alterations across genes and samples, a randomization procedure was applied to detect gene pairs with significant co-occurring or mutual exclusive alterations (see Materials and Methods; false discovery rate [FDR] < 0.05; Figure 1B). The statistics of co-occurring and mutual exclusive gene pairs within each cancer type are shown in Figure 2A.
Figure 2.
Identification of Candidate Synthetic Viable and Synthetic Lethal Interactions in Cancer
(A) Statistics of co-occurring and mutually exclusive gene pairs in different types of cancer. (B) Overlapping of the candidate synthetic viable interactions verified in the shRNA, CRISPR, and yeast datasets. (C) Overlapping of candidate synthetic lethal interactions verified in the shRNA, CRISPR, and yeast datasets.
The co-occurring and mutual exclusive gene pairs, which had a combined effect verified by at least one of the shRNA, CRISPR, and yeast datasets, were selected as candidate SV and SL interactions. According to the criteria to select candidate SV and SL interactions (see Materials and Methods), 15,928, 103,879, and 191 candidate SV interactions were separately identified in the shRNA, CRISPR, and yeast datasets (Figure 2B). In total, 11,001, 12,421 and 196 candidate SL interactions were separately identified in the shRNA, CRISPR, and yeast datasets (Figure 2C). The SL and SV interactions from 16 studies were integrated for follow-up research accompanied by our predictions. The information of 16 studies is provided in the Supplemental Materials and Methods. In total, 490 SV interactions and 29,171 SL interactions were selected from current studies (Table S1). An integrative confidence score combining scores from all evidence sources and our prediction were calculated to estimate the reliability of SL or SV genetic interaction (see Supplemental Materials and Methods). The quantitative score according to the experimental methods was annotated in evidence sources (Table S2).
SV Interactions and SL Interactions Associated with Drug Response
Four drug datasets (from CCLE, GDSC, Cancer Therapeutics Response Portal [CTRP], and NCI60) were used to identify drug biomarkers by investigating the SV and SL interactions in cancer cells. Our previous studies have demonstrated that drug response heterogeneity exists among tissue-specific cell lines.14 In the present study, there were significant differences in drug sensitivity among different tissues according to ANOVA (p < 0.05; Figure S1). Therefore, the drug responses of SV and SL interactions were analyzed in tissue-specific cell lines, which limited the number of cell lines to three. For SV interactions, if one gene was targeted by a particular drug, the drug response measures (IC50 or AUC) in cell lines with alterations of the partner gene were significantly higher than those in the cell lines with the wild-type (WT) partner gene. For SL interactions, if one gene targeted was by a particular drug, the drug response measures (IC50 or AUC) in cell lines with alterations of the partner gene were significantly lower than those in the cell lines with the WT partner gene. The SV and SL interactions conformed to the expected drug response were different for each tissue (Figures S2A and S2C). The significant overlapping of drug-response-related SV (or SL) interactions identified by AUC and LN_IC50 (natural log of the fitted half maximal inhibitory concentration) values from GDSC indicated the reliability of the present study (p < 0.001; Figures S2B and S2D). A well-known example of SL is that ovarian cancer patients with BRCA1/2 mutations are treated with PARP inhibitors. Figure S3 demonstrates that the BRCA2 mutation was associated with sensitivity to olaparib, a PARP inhibitor, in cancer cells.
Here, we calculated the percentage of the SV (SL) interactions that conformed to our hypothesis relative to all significant SV (SL) interactions and defined it as consistency ratio. The consistency ratios of four drugs (including erlotinib, lapatinib, nilotinib, and paclitaxel) were simultaneously screened in four drug datasets, which are shown in Figures 3A and 3B. In the NCI60 dataset, no SV and SL interactions exhibited the expected drug response due to the small number of cell lines. As shown in Figure 3A, the SV interactions associated with paclitaxel resistance had a higher consistency ratio in breast cancer, stomach cancer, and lung cancer. Overall, the response rate to paclitaxel is 50% as first-line chemotherapy, decreasing to 20%–30% when used as second- or third-line chemotherapy; nearly half of the patients with breast cancer do not respond to paclitaxel.15 According to the present results, the breast cancer cell lines with alterations of PSEN1 were related to paclitaxel resistance (Figure S4), which has been reported in esophageal cancer.16
Figure 3.
Genetic Interactions Related to Drug Response in Tissue-Specific Cell Lines
(A and B) Consistency ratio of (A) SV interactions related to drug resistance and (B) SL interactions related to drug sensitivity of four drugs (p < 0.05, Wilcoxon rank-sum test, white; and p < 0.01, Wilcoxon rank-sum test, black) in CCLE, CTRP, and GDSC datasets. Dark purple indicates a higher consistency ratio, and grids lacking purple indicate no detection. Different colors represent different tissues. The blank grid indicates that the consistency ratio cannot be tested because of a limited number of cell lines. (C) Consistency ratio of erlotinib in lung cancer tissue. (D–G) Cell lines with EGFR amplification or mutation are sensitive to the ERBB2-related drug, erlotinib, in CCLE, CTRP, and GDSC (AUC, LN_IC50) datasets (Wilcoxon rank-sum test). (D) IC50 of erlotinib in CCLE. (E) AUC of erlotinib in CTRP. (F) AUC of erlotinib in GDSC. (G) LN_IC50 of erlotinib in GDSC.
Erlotinib is an oral HER1/EGFR tyrosine kinase inhibitor approved for patients with non-small-cell lung cancer. In the present results, erlotinib showed a high consistency ratio in lung cancer tissue (Figure 3B), and the consistency ratio was 100% in the CCLE, GDSC (AUC), and GDSC (LN_IC50) datasets, p < 0.01 (Figure 3C). Cell lines with alterations of EGFR were more sensitive to erlotinib in the CCLE (Figure 3D), CTRP (Figure 3E), and GDSC (Figures 3F and 3G) datasets, which demonstrated the reliability of the present results. Together, these findings suggested that SV interactions are associated with drug resistance and that SL interactions are related to drug sensitivity.
Drug-Response-Related Genetic Interaction Network Shows Biological Characteristics
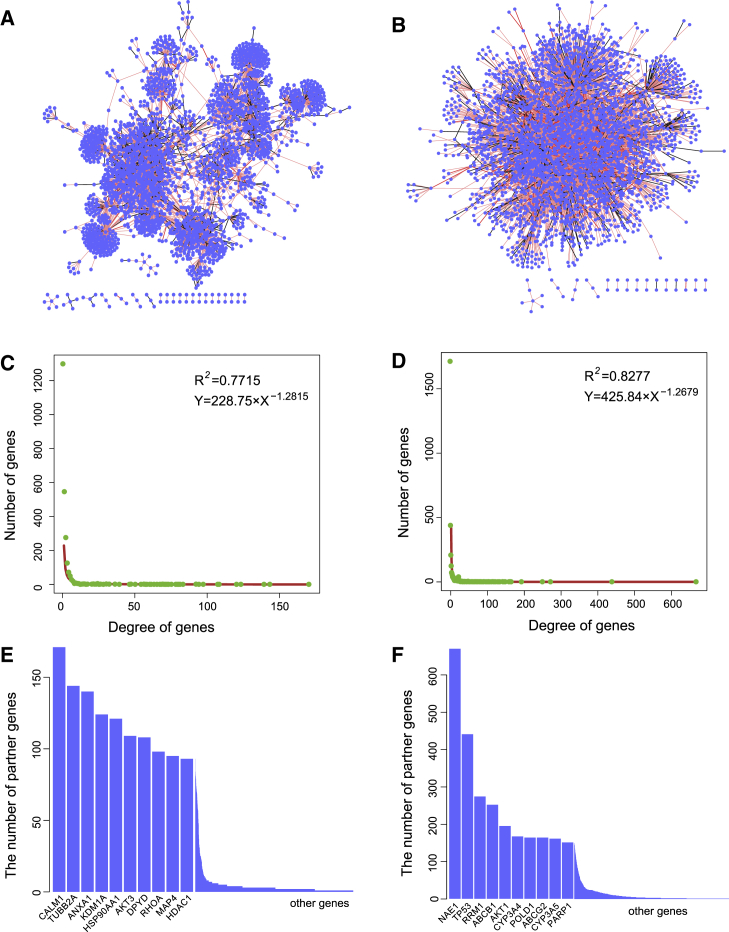
By integrating drug-resistance-related SV interactions and drug-sensitivity-related SL interactions, SV and SL networks, respectively, were constructed. The SV interaction network consisted of 2,516 genes and 4,529 target-partner gene relationships (Figure 4A), and the SL interaction network consisted of 3,184 genes and 10,637 target-partner gene relationships (Figure 4B); within two networks, 80% of SV interactions and 90% of SL interactions had one-step (red edge) or two-step (pink edge) neighbors in the protein-protein interaction (PPI) network, which was not expected by random chance (p < 0.001). Random SV and SL networks were constructed by randomly selecting the same number of gene pairs from the cancer alteration profiles and counting the number of one- or two-step interactions that overlapped with the PPI network, which was performed 1,000 times. Also, we calculated the empirical p value. Both SL and SV networks showed scale-free characteristics (Figures 4C and 4D). The aforementioned results indicated that the SV and SL networks had biological functions.17 The top 10 genes with the highest degrees in the SV and SL networks are shown in Figures 4E and 4F. Several drug targets, such as AKT1, PARP1, and HDAC1, had high degrees in the SV or SL network. Moreover, partner genes of these drug targets also had high degrees. For example, the RHOA partner gene in the SV network is a member of the Rho family of small GTPases, which functions as molecular switches in signal transduction cascades. The present study predicted that alterations of RHOA may induce resistance to drugs, such as fluorouracil, in cancer cells. Several studies have reported that alterations of RHOA mediate tumor invasion and drug resistance in cancer. Misek et al.18 found that RHOA contributes to BRAF inhibitor resistance in Sox9High/Sox10Low melanoma cells by regulating activation of the RhoA family signaling pathway. In addition, partner genes with high degrees in the SL network, such as NAE1 and TP53, also played an important role in the development and progression of cancer. The present work predicted that alterations of TP53 were related to drug sensitivity, which has been validated in other studies.19 Thus, the partner genes of drug targets in the SL and SV networks may be regarded as candidate biomarkers for cancer therapies.
Figure 4.
Analysis of Genetic Interaction Networks
(A) SV interaction network. (B) SL interaction network. Nodes depict genes, and edges represent SV interactions between genes. The light red line indicates that two genes with SV interactions also have direct protein-protein interactions, and the pink line indicates that two genes with SV or SL interactions have common neighbors in the protein-protein interaction network. (C) Distribution of the degree of genes in the SV network. (D) Distribution of the degree of genes in the SL network. (E) Histogram shows the degree of genes in the SV interaction network. (F) Histogram shows the degree of genes in the SL interaction network.
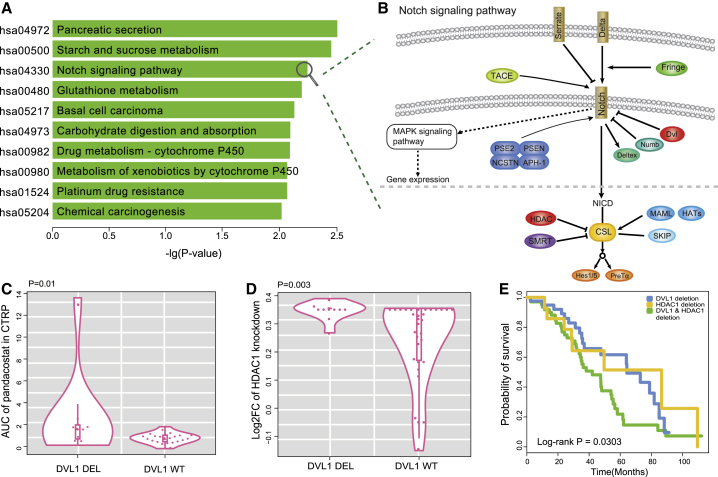
SV Interactions Induce Drug Resistance
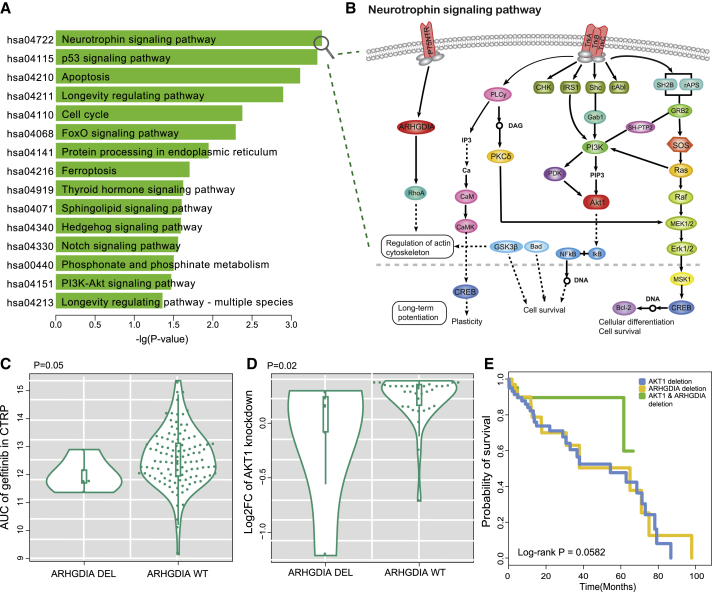
Among the genes with the top 10 highest degrees in the SV network, HDAC1 is a key drug target approved by the FDA. In the SV network, many genes interacted with HDAC1, whose alterations may induce resistance to a HDAC1 inhibitor in cancer cells. The partner genes of HDAC1 were significantly enriched in 35 pathways (hypergeometric test, p < 0.05; Figure S5), including Notch, Wnt, and mTOR signaling pathways (Figure 5A). Both HDAC1 and the DVL1 partner gene were important regulators in the Notch signaling pathway (Figure 5B), which governs the growth and proliferation of cancer cells. In the pharmacogenomic dataset of CTRP, the deletions of DVL1 were related to HDAC1 inhibitor (pandacostat) resistance in the cell lines of CNS tissues (p = 0.01, Wilcoxon rank-sum test; Figure 5C). Knockout of HDAC1 by CRISPR demonstrated that cell lines with DVL1 deletion survived better than those with WT DVL1 (p = 0.003, Wilcoxon rank-sum test; Figure 5D). HDAC1 is a member of the histone deacetylase/AcuC/AphA family whose main function is to remove acetyl from histone. HDAC1 is an inhibitor of the CSL protein in the Notch pathway, which can form a transcriptionally active complex with the Notch1 intracellular domain (NICD) to activate downstream targets of the transcriptional suppressor family, such as HES1/5 and HERP (Figure 5B). If HDAC1 is suppressed by pandacostat, transcriptional inhibition can be promoted. However, deletion of DVL1 may compensate for transcriptional inhibition. DVL1 inhibits the Notch1 receptor, and deletion of DVL1 may upregulate Notch1, which produces anti-apoptotic signals by regulating the phosphatidylinositol 3-kinase (PI3K)-PKB/Akt pathway.20 To confirm this conjecture, differentially expressed genes were examined in liver hepatocellular carcinoma (LIHC) samples containing both deletions of HDAC1 and DVL1 versus LIHC samples with WT HDAC1 and DVL1. Notch1 was significantly upregulated in samples containing deletions of both HDAC1 and DVL1 (p = 0.001, t test; Figure S6). Thus, when HDAC1 inhibitors promote transcriptional inhibition, the deletion of DVL1 may lead to an increase in the activity of the Notch receptor, which promotes cell proliferation by regulating the mitogen-activated protein kinase (MAPK) signaling pathway. To test whether this SV interaction affects patient survival, a log-rank test was applied to analyze the survival data of patients from TCGA. In ovarian cancer, patients with deletions of both HDAC1 and DVL1 showed worse survival than patients with deletions of either HDAC1 or DVL1 (p = 0.0303, log-rank test).
Figure 5.
Functional Analysis of Drug-Resistance-Related SV Interactions
(A) KEGG pathways enriched with the partner genes of HDAC1 in the SV interaction network. The y axis represents significantly enriched pathways, and the x axis is the negative log10-transformed hypergeometric test p value. (B) Notch signaling pathway. The pathway is composed of receptors, ligands, and the CSL DNA-binding protein. DVL1 is located in functional extracellular domains, acting as an inhibitor for Notch receptors, and HDAC1 plays a key role in regulating transcriptional activity by inhibiting CSL. (C) Deletions of DVL1 were related to pandacostat resistance in cell lines compared to wild-type DVL1 (p = 0.01, Wilcoxon rank-sum test). (D) Deletions of DVL1 showed higher viability in HDAC1 knockout cell lines compared to wild-type DVL1 (p = 0.003, Wilcoxon rank-sum test). (E) The Kaplan-Meier overall survival analysis of patients in three groups as follows: HDAC1 deletion, DVL1 deletion, and HDAC1 and DVL1 deletion (p = 0.0303, log-rank test).
SL Interactions Mediate Drug Sensitivity
AKT1, as a key gene encoding serine and threonine protein kinase in the PI3K/AKT pathway, interacted with gefitinib. Among genes that interacted with drugs in the SL network, AKT1 had the second largest number of partner genes (Figure S7). Pathway enrichment analysis showed that the partner genes of AKT1 were mainly enriched in the Neurotrophin signaling pathway, p53 signaling pathway, apoptosis pathway, and cell cycle (hypergeometric distribution, p < 0.05; Figures 6A and 6B). These results indicated that the alterations of the partner genes, which had an SL effect with deletion of AKT1, were related to gefitinib sensitivity in cancer cells. For example, ARHGDIA-deficient cell lines were related to gefitinib sensitivity in hematopoietic and lymphoid tissues (p = 0.05, Wilcoxon rank-sum test; Figure 6C). Knockout of AKT1 by CRISPR in cells with ARHGDIA deletion showed worse survival than cells with WT ARHGDIA (p = 0.02, Wilcoxon rank-sum test; Figure 6D). Both AKT1 and ARHGDIA participated in the Neurotrophin signaling pathway (Figure 6B), which regulates the growth, development, survival, and repair of the nervous system. AKT1, as the direct downstream target of PI3K, plays an important role in maintaining the survival of cerebellar cells.21 ARHGDIA, a downstream gene of p75NTR, has been proven to regulate the survival of neurons.22 Monje et al.23 confirmed that neuron-mediated developmental mechanisms are recapitulated in cancers. Cancer patient prognostic data were used to test whether co-deletions of AKT1 and ARHGDIA are beneficial to patient survival. As shown in Figure 6E, the prognosis of lung squamous cell carcinoma patients with AKT1 and ARHGDIA deletions was better than that for patients with single-gene deletions (p = 0.0582, log-rank test). Thus, alterations of ARHGDIA in cancer cells are beneficial for gefitinib treatment, and patients with co-alterations of AKT1 and ARHGDIA have better prognosis.
Figure 6.
Functional Analysis of Drug-Sensitivity-Related SL Interaction Genes
(A) KEGG pathways enriched with the partner genes of AKT1 in the SL interaction network. The y axis represents significantly enriched pathways, and the x axis represents the negative log10-transformed hypergeometric test p value. (B) Neurotrophin signaling pathway is mainly regulated by two types of receptors, the Trk tyrosine kinase receptors and the p75 neurotrophin receptor (p75NTR). AKT1 is the downstream effector of Trk, and ARHGDIA is the downstream gene of p75NTR. Both of these genes are important regulators for neuronal growth. (C) Deletions of ARHGDIA were related to gefitinib sensitivity in cell lines compared to wild-type ARHGDIA (p = 0.05, Wilcoxon rank-sum test). (D) Deletions of ARHGDIA showed lower viability in AKT1 knockout cell lines compared to wild-type ARHGDIA (p = 0.02, Wilcoxon rank-sum test). (E) The Kaplan-Meier overall survival (OS) analysis of patients in three groups as follows: AKT1 deletion, ARHGDIA deletion, and AKT1 and ARHGDIA deletion (p = 0.0582, log-rank test).
SL Interactions Suggest New Therapeutic Strategy
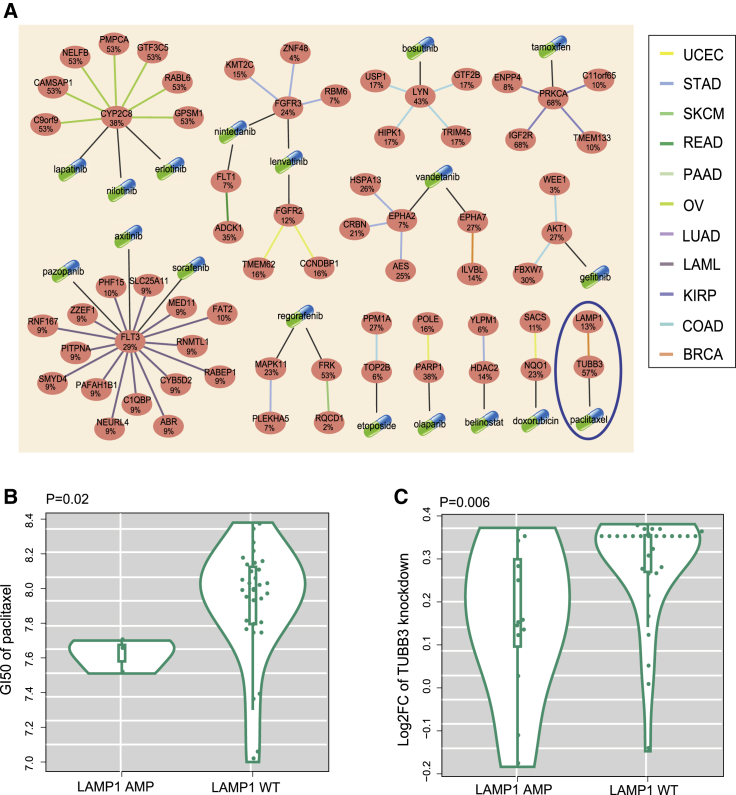
SL interactions provide a conceptual framework for the development of cancer-specific drugs.24 In the present results, alterations of partner genes involved in the SL network were related to drug sensitivity in tissue-specific cell lines, which indicated that drugs in the SL network may be new targeted therapeutics for specific cancer types (Figure 7A). To investigate whether drugs could be applied to specific cancers, the GI50 values for 90 drugs screened in breast cancer lines were evaluated.25 As expected, breast cancer cell lines with LAMP1 amplifications were related to paclitaxel sensitivity compared to WT cell lines (p = 0.02, Wilcoxon rank-sum test; Figure 7B). Knockout of TUBB3 by CRISPR in cells with LAMP1 amplifications showed worse survival than cells with WT LAMP1 (p = 0.006, Wilcoxon rank-sum test; Figure 7C). These results indicated that breast cancer patients with amplifications of LAMP1 may benefit from paclitaxel chemotherapy.
Figure 7.
SL Interactions Suggest New Therapeutic Strategy
(A) Network of potential anti-cancer therapies. Ellipse nodes represent genes, and the gene alteration rates in the specific cancer types are shown below the gene symbols. Capsules represent drugs. The black lines between capsules and ellipse node depict drug-target relationship. The edge colors represent the cancer types within which the synthetic lethal interactions were detected. (B) Breast cancer cell lines with LAMP1 amplifications had significantly lower GI50 of paclitaxel than breast cancer cell lines with wild-type LAMP1 (p = 0.02, Wilcoxon rank-sum test). (C) Knockout of TUBB3 by CRISPR in cancer cells with LAMP1 amplification showed better survival than cells with wild-type LAMP1 (p = 0.006, Wilcoxon rank-sum test).
Database of Genetic Interaction in Cancer
In the present work, we report that CancerGeneticInteractiondb (CGIdb) is a comprehensive platform for genetic interaction in cancer. In addition to providing candidate SV and SL interactions predicted by the present methods, CancerGeneticInteractiondb also collected a large number of SV and SL interactions predicted by other studies (Table S1). With a user-friendly interface, the website is composed of four sections: Search, Browse, Data, and About. On the Search page, the user can enter genes of interest and search for SV or SL interactions containing those genes. Furthermore, the drug response of SV (SL) interactions can be viewed by clicking the Detail button. In the Browse page, users can search SV and SL interactions by tissue types, and this page also provides drug information. Users can download and upload data on the Data page. For user convenience, all SV and SL interactions are grouped by tissues and data sources. Additional details for CancerGeneticInteractiondb are provided on the About page (also see Supplemental Materials and Methods). The website for accessing the database is located at http://www.medsysbio.org/CGIdb.
Discussion
The use of genetic data, pharmacological data, and functional screening datasets to gain valuable insights into the mechanism of drug sensitivity and drug resistance has been considered as the cornerstone of precision cancer medicine.26 Cancer cells have frequent defects in specific genes that drive growth and metastasis, and alterations in different genes may induce a genetic interaction effect. Identification of genetic interaction with cancer-promoting genes represents a compelling approach for the development of cancer therapies. The present study developed a new strategy for identifying genetic interactions in cancer cells based on CNAs, mutations, and expression profiles across 8,580 samples from 32 cancer types from the TCGA dataset, which were further filtered applying the shRNA, CRISPR, and yeast datasets. The present results demonstrated that SV interactions induce drug resistance and that SL interactions mediate drug sensitivity in cancer cells. The results highlighted that DVL1 deletion induces resistance to HDAC1 inhibitors through disturbing the Notch signaling pathway, which was confirmed by gene expression profiles. Within SL interactions, alterations of partner genes were related to drug sensitivity in cancer cell lines, suggesting that these genes may be new biomarkers for cancer therapies, thereby maximizing the clinical effectiveness of drugs. An expected application of genetic interaction screens is to optimize the therapeutic regimen of patients and to make an improvement in overall survival, which was verified in the clinical prognostic data. Finally, the present study provided a comprehensive CGIdb database for a convenient retrieval of genetic interactions in cancer.
It is well known that amplifications and deletions correspond to the upregulation and downregulation of gene expression, respectively. However, the functional effect of mutation on drug response is unclear. Somatic mutations can be classified as frameshift mutations, missense mutations, and so on. It is common for researchers to assume that frameshift insertions and deletions (indels) are loss-of-function variants.27 Missense mutations result in a single-nucleotide base change, which is difficult to use in predicting the effect on protein stability or PPI.28, 29 Investigating the functions of somatic mutation from structure, protein expression, and so on warrants our future work.
The different hypotheses of methods and original input data may result in low concordance of genetic interactions (Figures 2B and 2C). Ye et al.30 collected and compared the SL interactions identified by five studies. The results show that very little SL interactions are overlapped among different studies, which provided an indispensable reason that the genetic interaction relationships found in each work were only a small part because of the different hypotheses. Thus, beyond the predictions from our work, we collected results of 16 studies for follow-up research (Table S1).
Numerous studies have focused on identifying genetic interactions by shRNA or CRISPR technology, which provide large-scale functional screens in cell lines for genetic interaction detection.13, 31, 32, 33 CRISPR, a new technology of genome editing, has been widely applied to loss-of-function screens for essential genes, which have uncovered both core and cell-line-specific fitness genes. Large-scale CRISPR screens have been performed, and they provide new data sources for identifying genetic interactions.34, 35, 36 In the present study, the CRISPR screening dataset was added to further filter candidate SV and SL interactions. However, the shRNA or CRISPR studies were limited as follows: (1) lack of mutation information of cell lines—for example, the CCLE collection of 1,000 cell lines contained no acute myeloid leukemia (AML) cell line with IDH1 mutation, even though the rate of mutation in the AML patients is up to 10%;37 and (2) the inevitable difference of tumor microenvironment between in vitro screening conditions and in vivo tumors. The present method used human primary tumor data to identify potential cancer-specific SV and SL interactions to complement the limitations of existing cell-line screening methods.
The distribution of drug response values in cell lines derived from different tissues was significantly different. Therefore drug sensitivity in cancer cell lines is tissue specific (Figure S1). By the drug sensitivity analysis on the tissue-specific cell-line datasets of the large-scale pharmacogenomic data, most identified SL interactions with significant drug sensitivity conform with the SL hypothesis and most identified SV interactions with significant drug resistance conform with the SV hypothesis (Figure 3). However, the fewer cell-line samples for specific cancer types limited the statistical power. A new approach is urgently needed to estimate and remove the viability of drug sensitivity of different cells.
The purpose of the present work was to elucidate the mechanisms of drug resistance and drug sensitivity to exploit cancer-specific vulnerabilities and expand the scope of precision oncology. The partner genes of SL or SV identified in the present work had a close functional relationship with drug-interacted targets shown in Figures 4A and 4B. HDAC1, with the highest degree in the SV network (Figure 4A), was selected as a case study in the present work. HDAC1 is a transcription inhibitor and plays a key role in the regulation of eukaryotic gene expression accompanied by decreasing activity of transcription.38 Many studies have shown that HDAC1 may form a co-inhibitory complex with other chromatin regulatory proteins to regulate important biological processes, such as cell cycle and cell differentiation.39 At present, several HDAC inhibitors have been approved by the FDA for treatment of cancer. In the present results, several partner genes of HDAC1 induced drug resistance to HDAC1 inhibitors and participated in the same pathway with HDAC1. For example, DVL1 and HDAC1 participated in the Notch signaling pathway, which relies on the proteolytic cascade to release the transcriptional activity of the intracellular domain.40 When HDAC1 is inhibited by drugs to promote transcriptional inhibition in the Notch signaling pathway, DVL1 deletion activates the MAPK signaling pathway by the Notch receptor and further promotes cell proliferation. Thus, interactions between HDAC1 inhibition and DVL1 deletion may enhance cancer cell survival. Therefore, DVL1 deletion may induce resistance to HDAC1 inhibitors in cancer cells by disturbing the Notch signaling pathway, which was confirmed by the gene expression profile. NOTCH1 was significantly upregulated in samples with deletions of both HDAC1 and DVL1 compared to samples with single-gene deletions. Importantly, ovarian cancer patients with deletions of both HDAC1 and DVL1 had worse survival than patients with deletions of either HDAC1 or DVL1. Specific mechanisms were conjectured by bioinformatics statistics, but they should be verified by subsequent cell and animal experiments.
PARP1, as a well-known drug target, had high degrees in the SL network. PARP inhibitors are the first clinically approved drugs designed to exploit SL.8 Therefore, it would be meaningful to expand the application of PARP inhibitors in the clinic. The present analysis identified the well-known SL interaction between PARP inhibition and BRCA1 mutation. In the present results, alterations of the partner genes of PARP in the SL network were significantly related to the sensitivity of PARP inhibitors in cancer cell lines. For example, the mutation of POLE mediated sensitivity to the PARP inhibitor veliparib in lung cancer cell lines, which was previously characterized by Safiri et al.41 In addition, there were many newly discovered alterations of genes that enhanced cell-line sensitivity to PARP inhibitors, such as DOCK3, CLASP2, CMTM6, and MAP4. The new discoveries warrant additional detailed research, and experimental verification (cell and animal models) is required to unravel the mechanism of drug sensitivity, which will be the major focus in our future studies.
Materials and Methods
Identification of Candidate Genetic Interactions
The present study used mutation, CNA, and expression profiles derived across 32 cancer types from the TCGA consortium, including 8,580 patient samples. High-frequency somatic mutations were identified via the MutSig algorithm from whole-exome sequencing data.42 CNAs were determined by Genomic Identification of Significant Targets in Cancer (GISTIC).43 Similar to Shi et al.,44 the log2 ratio cutoff values >0.25 were defined as amplification. In contrast, the log2 ratio cutoff values < −0.25 were defined as deletion. To further optimize CNAs, Pearson’s correlation test was used to select genes whose copy number and expression value had a positive correlation coefficient. The p value was corrected using the Benjamini-Hochberg (BH) correction for multiple tests, and the results with an FDR < 0.05 were reported in the present study.
Integration of mutation and CNA data allowed generation of alteration profiles for 32 cancer types. The alteration profile columns represent samples, and the rows represent altered genes (Figure 1B). To discover gene pairs with significant co-occurrence or mutual exclusivity, a permutation strategy that controls for alteration heterogeneity within and across tumor samples was applied.3 Permutated genomic alteration matrices were implemented by the permatswap function in the R package vegan (http://vegan.r-forge.r-project.org/), which maintains the total number of alterations for each gene across samples as well as the total number of alterations per sample. In addition, due to the bias in alteration frequencies among cancer types, perturbations for different cancer samples were separately performed. The proportion of 10,000 permutations in which the observed co-occurrence was higher (Pco) or lower (Pme) than in the real data was taken as an empirical p value (Figure 1B). To avoid bias of limited number of permutation experiments, the hypergeometric test was used to further filter co-occurrence and mutually exclusive gene pairs. Bonferroni was used to correct the p value, and FDR < 0.001 was reported as significant. In addition, we have removed the co-occurring gene pairs from the same region of CNAs.
shRNA-Based Screening
shRNA data were downloaded from The Project Achilles database, which provides shRNA depletion scores from pooled genomic library tests across 216 cancer cell lines. A gene-level composite score (shRNA score) was obtained by Analytic Technique for Assessment of RNAi by Similarity (ATARiS) (https://www.broadinstitute.org/achilles/datasets/all). Higher shRNA scores indicated enhancement of cell viability. The genetic background of these cell lines was obtained from the Cancer Cell Line Encyclopedia (CCLE; https://www.broadinstitute.org/ccle/home), including mutation and copy number variation. Genomic alteration profiles of cell lines were built. In the genomic alteration profile, the row represented the gene, and the column represented the cell line. For each pair of genes (G1 and G2), shRNA scores of G1 knockdown were compared between the cell lines with and without G2 alterations using one-sided Wilcoxon rank-sum test (Figure 1C).
CRISPR-Based Screening
High-throughput CRISPR-Cas9 screening data, including 63 cancer cell lines, were downloaded from the GenomeCRISPR database. To compare fitness phenotypes of CRISPR-Cas9 screens, all screens were reanalyzed through the Bayesian Analysis of Gene Essentiality (BAGEL).45 The Bayesian factor serves as a quantitative measure of the essentiality of the gene in the screen, in which higher Bayesian factors indicate more essential genes. Negative Bayesian factors were used as fitness scores. A lower fitness score represents higher confidence that a given gene knockout causes a decrease in fitness.17 CNA and mutation data of cell lines were obtained from CCLE. In cell lines with knockout of G1, a one-sided Wilcoxon rank-sum test was used to test whether the fitness scores were significantly higher or lower in cell lines with and without alterations of G2 (Figure 1C).
Yeast-Based Comparative Genomic Strategy
A genome-scale genetic interaction map in yeast was obtained from Costanzo et al.,46 who generated quantitative genetic interaction profiles for approximately 75% of all genes in budding yeast. Deshpande et al.47 developed a comparative genomic algorithm to identify cancer-related genetic interactions. The present study used a cutoff of ε > 0.08 with p < 0.05 (yeast p value) to detect yeast SV interactions and ε < −0.08 with p < 0.05 to detect yeast SL interactions (ε is a measure of genetic interaction strength) (Figure 1C). A positive ε represents an increase in cell fitness, and a negative ε represents a decrease in cell fitness. InParanoid7 was applied to map yeast genes to human genes. Only 1:1 orthologs were used in the present study.
Lastly, the co-occurrence and mutual exclusivity gene pairs verified in at least one type of the aforementioned three datasets were selected as candidate genetic interactions (Figure 1C).
Identifying SV and SL Interactions Related to Drug Response
The original pharmacological screening data were downloaded from CCLE,48 Genomics of Drug Sensitivity in Cancer (GDSC),49 Broad CTRP (https://www.broadinstitute.org/ctrp/), and NCI60.50, 51 The CCLE dataset contained IC50 values of 24 anti-cancer compounds across 501 cancer cell lines derived from 23 cancer types. The GDSC data contained IC50 values and the area under the drug inhibition curve (AUC) of 251 drugs detected in 1,012 cell lines from 30 cancer types. The CTRP data contained the AUC of 481 small molecules used to treat 823 cancer cell lines from 23 cancer types. The NCI60 data contained the concentration required for cell growth inhibition by 50% (GI50) of 4,329 anti-cancer compounds across 60 cancer cell lines. The information of drug and targeted genes or affected genes was collected from Drug Bank,52 ChEMBL,53 CCLE, and Catalogue of Somatic Mutations in Cancer (COSMIC).54
For a G1 and G2 gene pair, the IC50 (or AUC or GI50) of cell lines carrying G2 alterations and cell lines containing WT G2 were compared using a one-sided Wilcoxon rank-sum test if G1 in the SV pair was targeted by a drug. Only cancer types with more than 3 cell lines were included for statistical analyses.
Generation of Genetic Interaction Network and Functional Analysis
The network of SV related to drug resistance and the network of SL related to drug sensitivity were constructed. Cytoscape software was used to visualize the networks (https://cytoscape.org/). PPI data was downloaded from the Pathway Commons database.55 A hypergeometric distribution model was used to test whether the partner genes in the SV (SL) network were significantly enriched in biological pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.56
Prognosis Analysis
The mutation and CNA profiles from TCGA were analyzed to examine the prognostic value embedded in the SV and SL networks. The patients were divided into three groups according to the status of genes in the SV (SL) interactions as follows: G1 alteration only, G2 alteration only, and G1 and G2 alteration groups. The overall survival time of the three patient groups for specific cancer types was tested by using log-rank test, and the results were represented by Kaplan-Meier plots.
CGIdb Construction
CGIdb is freely available at http://www.medsysbio.org/CGIdb with no registration or login. CGIdb was implemented by HTML and Django, which is a high-level Python web framework. The dynamic backend of CGIdb was realized through the Linux, Apache, MySQL, and Python (LAMP) architecture. The interface component was designed using the Cascading Styling Sheet (CSS) and JavaScript, which have been tested in Google Chrome and Firefox browsers.
Author Contributions
Y.G. and Z.G. guided the research study. Y.H., C.W., and Q.D. completed data processing, analysis, and draft of the paper. C.W. and Z.Z. constructed the database. T.C., F.Y., Y.L., and B.C. extracted data and visualized the results. L.Q., W.Z., and H.L. provided valuable suggestions for the manuscript.
Acknowledgments
The authors acknowledge the efforts of all of the researchers who have contributed the data to the public databases of CCLE, CTRP, GDSC, NCI60, TCGA, SynLethDB, and METABRIC. The interpretation and reporting of these data are the sole responsibility of the authors. This work was supported by the National Natural Science Foundation of China (nos. 61673143, 81572935, 81872396, 61701143, and 61601151); the Postdoctoral Scientific Research Developmental Fund (no. LBH-Q16166); and the University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (no. UNPYSCT-2017061).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.07.003.
Contributor Information
Zheng Guo, Email: guoz@ems.hrbmu.edu.cn.
Yunyan Gu, Email: guyunyan@ems.hrbmu.edu.cn.
Supplemental Information
References
- 1.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Santhosh S., Kumar P., Ramprasad V., Chaudhuri A. Evolution of targeted therapies in cancer: opportunities and challenges in the clinic. Future Oncol. 2015;11:279–293. doi: 10.2217/fon.14.198. [DOI] [PubMed] [Google Scholar]
- 3.Park S., Lehner B. Cancer type-dependent genetic interactions between cancer driver alterations indicate plasticity of epistasis across cell types. Mol. Syst. Biol. 2015;11:824. doi: 10.15252/msb.20156102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinha S., Thomas D., Chan S., Gao Y., Brunen D., Torabi D., Reinisch A., Hernandez D., Chan A., Rankin E.B. Systematic discovery of mutation-specific synthetic lethals by mining pan-cancer human primary tumor data. Nat. Commun. 2017;8:15580. doi: 10.1038/ncomms15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu Y., Wang R., Han Y., Zhou W., Zhao Z., Chen T., Zhang Y., Peng F., Liang H., Qi L. A landscape of synthetic viable interactions in cancer. Brief. Bioinform. 2018;19:644–655. doi: 10.1093/bib/bbw142. [DOI] [PubMed] [Google Scholar]
- 6.Ashworth A., Lord C.J., Reis-Filho J.S. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 7.Li X.J., Mishra S.K., Wu M., Zhang F., Zheng J. Syn-lethality: an integrative knowledge base of synthetic lethality towards discovery of selective anticancer therapies. Biomed Res. Int. 2014;2014:196034. doi: 10.1155/2014/196034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lord C.J., Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin K.K., Harrell M.I., Oza A.M., Oaknin A., Ray-Coquard I., Tinker A.V., Helman E., Radke M.R., Say C., Vo L.T. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2019;9:210–219. doi: 10.1158/2159-8290.CD-18-0715. [DOI] [PubMed] [Google Scholar]
- 10.Hu H.M., Zhao X., Kaushik S., Robillard L., Barthelet A., Lin K.K., Shah K.N., Simmons A.D., Raponi M., Harding T.C., Bandyopadhyay S. A quantitative chemotherapy genetic interaction map reveals factors associated with PARP inhibitor resistance. Cell Rep. 2018;23:918–929. doi: 10.1016/j.celrep.2018.03.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochoa S., Martinez-Perez E., Zea D.J., Molina-Vila M.A., Marino-Buslje C. Co-mutation and exclusion analysis in human tumors, a means for cancer biology studies and treatment design. bioRxiv. 2017 doi: 10.1101/182501. [DOI] [PubMed] [Google Scholar]
- 12.Unni A.M., Lockwood W.W., Zejnullahu K., Lee-Lin S.Q., Varmus H. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. eLife. 2015;4:e06907. doi: 10.7554/eLife.06907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson J.M., Nguyen Q.H., Singh M., Razorenova O.V. Approaches to identifying synthetic lethal interactions in cancer. Yale J. Biol. Med. 2015;88:145–155. [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R., Han Y., Zhao Z., Yang F., Chen T., Zhou W., Wang X., Qi L., Zhao W., Guo Z., Gu Y. Link synthetic lethality to drug sensitivity of cancer cells. Brief. Bioinform. 2017 doi: 10.1093/bib/bbx172. Published online December 28, 2017. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Y.Z., Yu K.D., Peng W.T., Di G.H., Wu J., Liu G.Y., Shao Z.M. Enriched variations in TEKT4 and breast cancer resistance to paclitaxel. Nat. Commun. 2014;5:3802. doi: 10.1038/ncomms4802. [DOI] [PubMed] [Google Scholar]
- 16.Meng F., Qian L., Lv L., Ding B., Zhou G., Cheng X., Niu S., Liang Y. miR-193a-3p regulation of chemoradiation resistance in oesophageal cancer cells via the PSEN1 gene. Gene. 2016;579:139–145. doi: 10.1016/j.gene.2015.12.060. [DOI] [PubMed] [Google Scholar]
- 17.Rauscher B., Heigwer F., Henkel L., Hielscher T., Voloshanenko O., Boutros M. Toward an integrated map of genetic interactions in cancer cells. Mol. Syst. Biol. 2018;14:e7656. doi: 10.15252/msb.20177656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misek S.A., Appleton K.M., Dexheimer T.S., Lisabeth E.M., Lo R., Larsen S.D., Gallo K., Neubig R.R. Rho-mediated MRTF and YAP1 activation contributes to BRAF inhibitor resistance in Sox9High/Sox10Low melanoma cells. bioRxiv. 2018 doi: 10.1101/381806. [DOI] [Google Scholar]
- 19.Liu Y., Zhang X., Han C., Wan G., Huang X., Ivan C., Jiang D., Rodriguez-Aguayo C., Lopez-Berestein G., Rao P.H. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015;520:697–701. doi: 10.1038/nature14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nair P., Somasundaram K., Krishna S. Activated Notch1 inhibits p53-induced apoptosis and sustains transformation by human papillomavirus type 16 E6 and E7 oncogenes through a PI3K-PKB/Akt-dependent pathway. J. Virol. 2003;77:7106–7112. doi: 10.1128/JVI.77.12.7106-7112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao M.V. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat. Rev. Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- 22.Ciavardelli D., Silvestri E., Del Viscovo A., Bomba M., De Gregorio D., Moreno M., Di Ilio C., Goglia F., Canzoniero L.M., Sensi S.L. Alterations of brain and cerebellar proteomes linked to Aβ and tau pathology in a female triple-transgenic murine model of Alzheimer’s disease. Cell Death Dis. 2010;1:e90. doi: 10.1038/cddis.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkatesh H., Monje M. Neuronal activity in ontogeny and oncology. Trends Cancer. 2017;3:89–112. doi: 10.1016/j.trecan.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinhardt H.C., Jiang H., Hemann M.T., Yaffe M.B. Exploiting synthetic lethal interactions for targeted cancer therapy. Cell Cycle. 2009;8:3112–3119. doi: 10.4161/cc.8.19.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daemen A., Griffith O.L., Heiser L.M., Wang N.J., Enache O.M., Sanborn Z., Pepin F., Durinck S., Korkola J.E., Griffith M. Modeling precision treatment of breast cancer. Genome Biol. 2013;14:R110. doi: 10.1186/gb-2013-14-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berlow N., Haider S., Wan Q., Geltzeiler M., Davis L.E., Keller C., Pal R. An integrated approach to anti-cancer drug sensitivity prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014;11:995–1008. doi: 10.1109/TCBB.2014.2321138. [DOI] [PubMed] [Google Scholar]
- 27.Hu J., Ng P.C. Predicting the effects of frameshifting indels. Genome Biol. 2012;13:R9. doi: 10.1186/gb-2012-13-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandurangan A.P., Ochoa-Montaño B., Ascher D.B., Blundell T.L. SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017;45(W1):W229–W235. doi: 10.1093/nar/gkx439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brender J.R., Zhang Y. Predicting the effect of mutations on protein-protein binding interactions through structure-based interface profiles. PLoS Comput. Biol. 2015;11:e1004494. doi: 10.1371/journal.pcbi.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye H., Zhang X., Chen Y., Liu Q., Wei J. Ranking novel cancer driving synthetic lethal gene pairs using TCGA data. Oncotarget. 2016;7:55352–55367. doi: 10.18632/oncotarget.10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vizeacoumar F.J., Arnold R., Vizeacoumar F.S., Chandrashekhar M., Buzina A., Young J.T., Kwan J.H., Sayad A., Mero P., Lawo S. A negative genetic interaction map in isogenic cancer cell lines reveals cancer cell vulnerabilities. Mol. Syst. Biol. 2013;9:696. doi: 10.1038/msb.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T., Yu H., Hughes N.W., Liu B., Kendirli A., Klein K., Chen W.W., Lander E.S., Sabatini D.M. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 2017;168:890–903.e15. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han K., Jeng E.E., Hess G.T., Morgens D.W., Li A., Bassik M.C. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 2017;35:463–474. doi: 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen J.P., Zhao D., Sasik R., Luebeck J., Birmingham A., Bojorquez-Gomez A., Licon K., Klepper K., Pekin D., Beckett A.N. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat. Methods. 2017;14:573–576. doi: 10.1038/nmeth.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertomeu T., Coulombe-Huntington J., Chatr-Aryamontri A., Bourdages K.G., Coyaud E., Raught B., Xia Y., Tyers M. A high-resolution genome-wide CRISPR/Cas9 viability screen reveals structural features and contextual diversity of the human cell-essential proteome. Mol. Cell. Biol. 2017;38 doi: 10.1128/MCB.00302-17. 00302-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W., Baty J.D., Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta M., Staszewski O., Raschi E., Frosch M., Hagemeyer N., Tay T.L., Blank T., Kreutzfeldt M., Merkler D., Ziegler-Waldkirch S. Histone deacetylases 1 and 2 regulate microglia function during development, homeostasis, and neurodegeneration in a context-dependent manner. Immunity. 2018;48:514–529.e6. doi: 10.1016/j.immuni.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Micheli L., D’Andrea G., Leonardi L., Tirone F. HDAC1, HDAC4, and HDAC9 bind to PC3/Tis21/Btg2 and are required for its inhibition of cell cycle progression and cyclin D1 expression. J. Cell. Physiol. 2017;232:1696–1707. doi: 10.1002/jcp.25467. [DOI] [PubMed] [Google Scholar]
- 40.Kovall R.A., Gebelein B., Sprinzak D., Kopan R. The canonical Notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev. Cell. 2017;41:228–241. doi: 10.1016/j.devcel.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Safiri S., Ayubi E. Smoking history predicts sensitivity to PARP inhibitor veliparib in patients with advanced non-small cell lung cancer: methodological issues. J. Thorac. Oncol. 2017;12:e130–e131. doi: 10.1016/j.jtho.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Forrest W.F., Cavet G. Comment on “The consensus coding sequences of human breast and colorectal cancers.”. Science. 2007;317:1500. doi: 10.1126/science.1138179. [DOI] [PubMed] [Google Scholar]
- 43.Beroukhim R., Getz G., Nghiemphu L., Barretina J., Hsueh T., Linhart D., Vivanco I., Lee J.C., Huang J.H., Alexander S. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Z.-Z., Jiang Y.-Y., Hao J.-J., Zhang Y., Zhang T.-T., Shang L., Liu S.-G., Shi F., Wang M.-R. Identification of putative target genes for amplification within 11q13.2 and 3q27.1 in esophageal squamous cell carcinoma. Clin. Transl. Oncol. 2014;16:606–615. doi: 10.1007/s12094-013-1124-z. [DOI] [PubMed] [Google Scholar]
- 45.Hart T., Moffat J. BAGEL: a computational framework for identifying essential genes from pooled library screens. BMC Bioinformatics. 2016;17:164. doi: 10.1186/s12859-016-1015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshpande R., Asiedu M.K., Klebig M., Sutor S., Kuzmin E., Nelson J., Piotrowski J., Shin S.H., Yoshida M., Costanzo M. A comparative genomic approach for identifying synthetic lethal interactions in human cancer. Cancer Res. 2013;73:6128–6136. doi: 10.1158/0008-5472.CAN-12-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rees M.G., Seashore-Ludlow B., Cheah J.H., Adams D.J., Price E.V., Gill S., Javaid S., Coletti M.E., Jones V.L., Bodycombe N.E. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016;12:109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shankavaram U.T., Varma S., Kane D., Sunshine M., Chary K.K., Reinhold W.C., Pommier Y., Weinstein J.N. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics. 2009;10:277. doi: 10.1186/1471-2164-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knox C., Law V., Jewison T., Liu P., Ly S., Frolkis A., Pon A., Banco K., Mak C., Neveu V. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., Overington J.P. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cerami E.G., Gross B.E., Demir E., Rodchenkov I., Babur O., Anwar N., Schultz N., Bader G.D., Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.