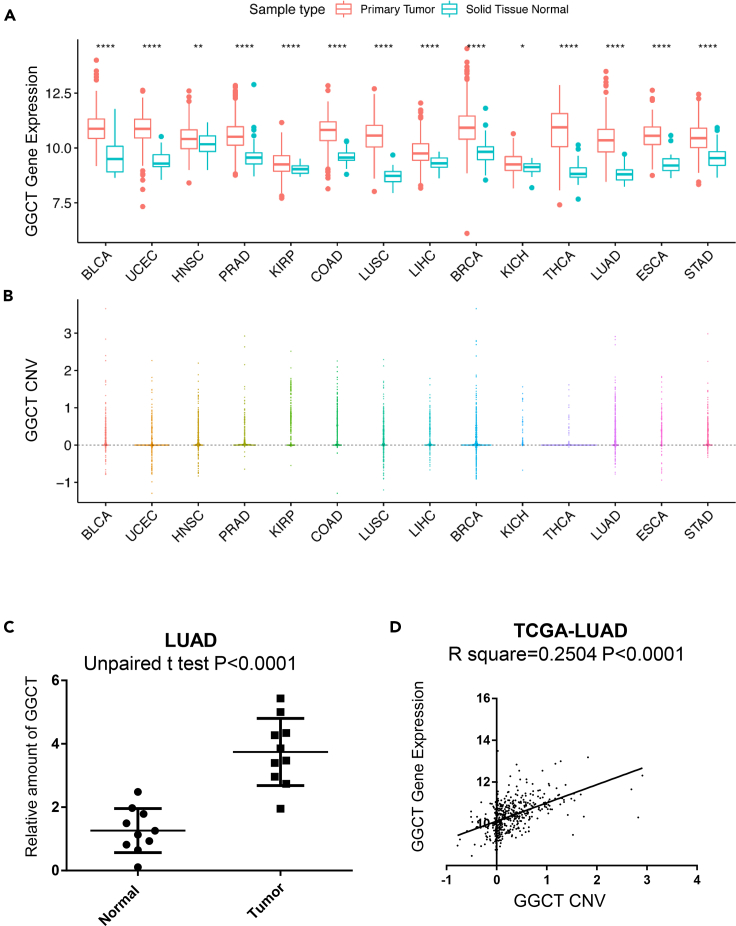
Figure 2.
GGCT Copy Number Variation (CNV) and mRNA Expression Status in Human Cancers
(A) GGCT mRNA expression levels (log2 based) were statistically up-regulated (unpaired two-tailed t test) in 14 of 15 types of human cancers when compared with corresponding normal control tissues based on The Cancer Genome Atlas (TCGA) database. Only 15 of 32 TCGA cancer types have both tumor and normal control samples available, and the number of normal samples is greater than or equal to 10. ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05.
(B) GGCT copy number values (log2 based ratio, normal copy number is 0) obtained from GISTIC2 software in cancers as (A) are shown based on TCGA database. GISTIC2 CNV value 0 means normal copy number.
(C) GGCT CNV values were detected by qPCR in patients with LUAD (n = 10) and normal control (n = 10) samples. Error bars represent mean ± SD. p Values of unpaired two-tailed t test are shown.
(D) The correlation between GGCT CNV and mRNA in TCGA lung adenocarcinoma (LUAD) samples (n = 511).