Abstract
Bacteria sense temporal changes in extracellular stimuli via sensory signal transducers and move by rotating flagella towards into a favorable environment for their survival. Each flagellum is a supramolecular motility machine consisting of a bi-directional rotary motor, a universal joint and a helical propeller. The signal transducers transmit environmental signals to the flagellar motor through a cytoplasmic chemotactic signaling pathway. The flagellar motor is composed of a rotor and multiple stator units, each of which acts as a transmembrane proton channel to conduct protons and exert force on the rotor. FliG, FliM and FliN form the C ring on the cytoplasmic face of the basal body MS ring made of the transmembrane protein FliF and act as the rotor. The C ring also serves as a switching device that enables the motor to spin in both counterclockwise (CCW) and clockwise (CW) directions. The phosphorylated form of the chemotactic signaling protein CheY binds to FliM and FliN to induce conformational changes of the C ring responsible for switching the direction of flagellar motor rotation from CCW to CW. In this mini-review, we will describe current understanding of the switching mechanism of the bacterial flagellar motor.
Keywords: Adaptive remodeling, Bacterial flagellar motor, Chemotaxis, Cooperativity, Directional switching, Motility
1. Introduction
Many bacteria possess flagella to swim in liquid media and move on solid surfaces. Escherichia coli and Salmonella enterica serovar Typhimurium (hereafter referred to Salmonella) are model organisms that have provided deep insights into the structure and function of the bacterial flagellum. The flagellum is composed of basal body rings and an axial structure consisting of at least three parts: the rod as a drive shaft, the hook as a universal joint and the filament as a helical propeller (Fig. 1A). The flagellar motor of E. coli and Salmonella consists of a rotor and a dozen stator units and is powered by an electrochemical potential of protons across the cytoplasmic membrane, namely proton motive force. Marine Vibrio and extremely alkalophilic Bacillus utilize sodium motive force as the energy source to drive flagellar motor rotation. The rotor is composed of the MS ring made of the transmembrane protein FliF and the C ring consisting of three cytoplasmic proteins, FliG, FliM and FliN. Each stator unit is composed of two transmembrane proteins, MotA and MotB, and acts as a transmembrane proton channel to couple the proton flow through the channel with torque generation (Fig. 1B) [[1], [2], [3], [4], [5]].
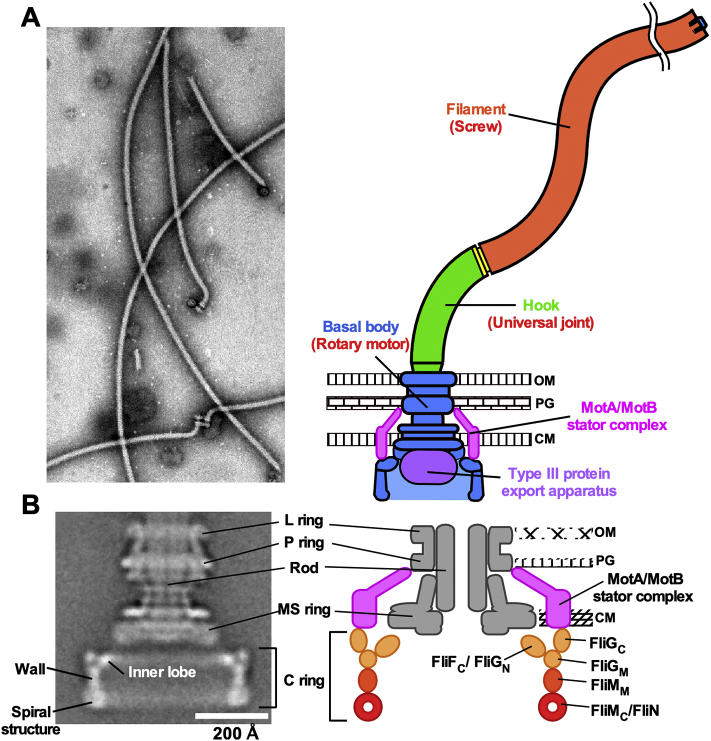
Fig. 1.
Subunit organization in the flagellar motor. (A) Bacterial flagella. Electron micrograph of flagella purified from Salmonella on the left and its schematic diagram on the right. The flagellum is composed of the bsal body as a rotary motor, the hook as a universal joint and the filament as a molecular screw. (B) CryoEM image of Salmonella basal body on the left and its schematic diagram on the right. The purified basal body consists of the C, MS, L and P rings and the rod. A dozen MotA/MotB stator complexes are associated with the basal body but are lost during purification. The C ring is composed of FliG, FliM and FliN. The N-terminal domain of FliG (FliGN) forms the inner lobe along with the C-terminal cytoplasmic domain of FliF (FliFC). The C-terminal domain of FliG (FliGC) is located in the upper part of the C ring wall. The middle domain of FliM (FliMM) is located between the middle domain of FliG (FliGM) and FliN and forms a cylindrical wall of the C ring. A continuous spiral density at the bottom edge of the C ring is made of the C-terminal domains of FliM (FliMC) and FliN.
The flagellar motor rotates in either counterclockwise (CCW; viewed from the flagellar filament to the motor) or clockwise (CW) direction in E. coli and Salmonella. When all the motors rotate in the CCW direction, flagellar filaments together form a bundle behind the cell body to push the cell forward. Brief CW rotation of one or more flagellar motors disrupts the flagellar bundle, allowing the cell to tumble, followed by a change in the swimming direction. Sensory signal transducers sense temporal changes in extracellular stimuli such as chemicals, temperature and pH and transmit such extracellular signals to the flagellar motor via the intracellular chemotactic signaling network. The phosphorylated form of CheY (CheY-P), which serves as a signaling molecule, binds to FliM and FliN in the C ring to switch the direction of flagellar motor rotation from CCW to CW. Thus, the C ring acts as a switching device to switch between the CCW and CW states of the motor [2,5].
The stator complex is composed of four copies of MotA and two copies of MotB. The MotA4/MotB2 complex is anchored to the peptidoglycan (PG) layer through direct interactions of the C-terminal periplasmic domain of MotB with the PG layer to become an active stator unit around the rotor [4]. A highly conserved aspartate residue of MotB (Asp-32 in the E. coli protein and Asp-33 in the Salmonella protein) is located in the MotA4/MotB2 proton channel and is involved in the energy coupling mechanism [6,7]. The cytoplasmic loop between transmembrane helices 2 and 3 of MotA (MotAC) contains highly conserved Arg-90 and Glu-98 residues and are important not only for torque generation but also for stator assembly around the rotor [[8], [9], [10]].
FliG is directly involved in torque generation [8]. Highly conserved Arg-281 and Asp-289 residues are located on the torque helix of FliG (HelixTorque) [11] and interact with Glu-98 and Arg-90 of MotAC, respectively [8,10]. Since the elementary process of torque generation caused by sequential stator–rotor interactions in the flagellar motor is symmetric in the CCW and CW rotation, HelixTorque is postulated to rotate 180° relative to MotAC in a highly cooperative manner when the motor switches between the CCW and CW states of the C ring [12]. This mini-review article covers current understanding of how such a cooperative remodeling of the C ring structure occurs.
2. Structure of the C Ring
FliF assembles into the MS ring within the cytoplasmic membrane [13]. The C ring consisting of a cylindrical wall and inner lobes is formed by FliG, FliM and FliN on the cytoplasmic face of the MS ring with the inner lobes connected to the MS ring (Fig. 1B) [14]. FliF requires FliG to facilitate MS ring formation in the cytoplasmic membrane [15]. FliG binds to FliF with a one-to-one stoichiometry [16]. FliM and FliN together form the FliM1/FliN3 complex consisting of one copy of FliM and three copies of FliN [17], and the FliM1/FliN3 complex binds to the FliG ring structure through a one-to-one interaction between FliG and FliM to form the continuous C ring wall [[18], [19], [20]]. Most of the domain structures of FliG, FliM and FliN have been solved at atomic resolution (Fig. 2), and possible models of their organization in the C ring have been proposed (Fig. 1B) [21,22].
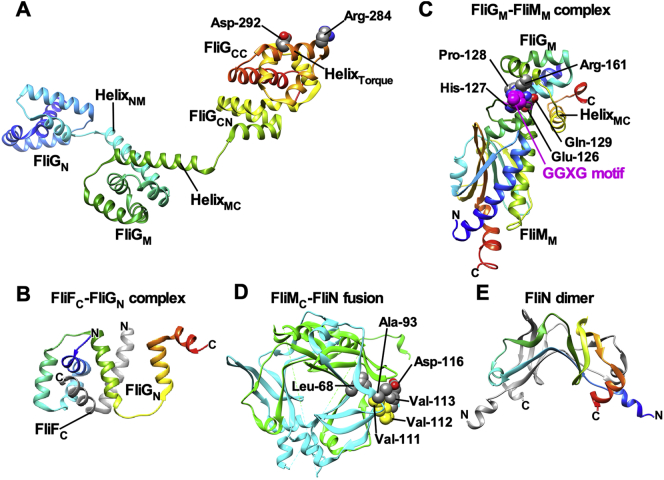
Fig. 2.
Crystal structures of C ring proteins. (A) Crystal structure of FliG derived from Aquifex aeolicus (PDB code: 3HJL). The Cα backbone is colour-coded from blue to red, going through the rainbow colors from the N- to the C-terminus. FliG consists of FliGN, FliGM and FliGC domains and two helix linkers, HelixNM and HelixMC. FliGC is divided into FliGCN and FliGCC subdomains. Arg-284 and Asp-292 residues, which correspond to Arg-281 and Asp-289 of E. coli FliG, respectively, are located in the torque helix of FliGCC (HelixTorque), which is involved in electrostatic interactions with the cytoplasmic loop of MotA. (B) Crystal structure of the FliFC/FliGN complex derived from Thermotaoga maritima (PDB code: 5TDY). FliFC consisting of two α-helices (grey) binds to a hydrophobic groove of FliGN (rainbow). (C) Crystal structure of the FliGM/FliMM complex derived from T. maritima (PDB code: 3SOH). A well conserved EHPQR motif in FliGM and a well conserved GGXG motif in FliMM are responsible for the FliGM–FliMM interaction. (D) Crystal structure of Salmonella FliMC-FliNN fusion protein (PDB code: 4YXB). FliMC and FliNN subunits are shown in green and cyan, respectively. Leu-68, Ala-93, Val-113 and Asp-116 of FliN are involved in the interaction with CheY-P. Val-111, Val-112 and Val-113 of FliN are required for the interaction with FliH. (E) Crystal structure of the FliN dimer derived from T. maritima (PDB code: 1YAB). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.1. FliG
FliG consists of three domains: N-terminal (FliGN), middle (FliGM) and C-terminal (FliGC) domains (Fig. 2A) [23]. FliGC is divided into two subdomains: FliGCN and FliGCC. FliGN is involved in the interaction with the C-terminal cytoplasmic domain of FliF (FliFC) (Fig. 2B) [24,25]. Inter-molecular interactions between FliGN and FliGN and between FliGM and FliGCN are responsible for the assembly of FliG into the ring structure on the cytoplasmic face of the MS ring [[26], [27], [28], [29]]. FliGM provides binding sites for FliM (Fig. 2C) [[18], [19], [20]]. A highly conserved EHPQR motif of FliGM is involved in the interaction with FliM [18,30]. FliGCC contains HelixTorque, and highly conserved Arg-284 and Asp-292 residues of Aquifex aeolicus FliG, which corresponds to Arg-281 and Asp-289 of E. coli FliG involved in the interactions with conserved charged residues of MotAC [8,11], are exposed to solvent on the surface of HelixTorque [23].
2.2. FliM
FliM consists of three domains: N-terminal (FliMN), middle (FliMM) and C-terminal (FliMC) domains [31,32]. FliMN contains a well conserved LSQXEIDALL sequence, which is responsible for the interaction with CheY-P [33]. FliMN is intrinsically disordered, and the binding of CheY-P to FliMN allows FliMN to become structured [32]. FliMM has a compactly folded conformation (Fig. 2C), and side-by-side associations between FliMM domains are responsible for the formation of the C ring wall [32]. The binding of CheY-P to FliMN affects inter-molecular FliMM–FliMM interactions, thereby inducing a conformational change in the C ring responsible for switching the direction of flagellar motor rotation [34]. A well conserved GGXG motif of FliMM is involved in the interaction with FliGM (Fig. 2C) [18,30]. FliMC shows significant sequence and structural similarities with FliN and is responsible for the interaction with FliN (Fig. 2D) [35].
2.3. FliN
FliN is composed of an intrinsically disordered N-terminal region (FliNN) and a compactly folded domain (FliNC), which structurally looks similar to FliMC [36]. FliN exists as a dimer of dimer in solution (Fig. 2E) [37] and forms the FliM1/FliN3 complex along with FliM through an interaction between FliMC and FliN [17]. CheY-P binds to FliNC in a FliM-dependent manner [38]. Leu-68, Ala-93, Val-113 and Asp-116 of E. coli FliN are responsible for the interaction with CheY-P (Fig. 2D) [38,39]. The binding of CheY-P to FliN affects interactions between FliMC and FliN, inducing the conformational change of the C ring responsible for directional switching of flagellar motor rotation [38]. FliNN seems to control the binding affinity of FliNC for CheY-P [38] although it is dispensable for the function of FliN [40]. FliN also provides binding sites for FliH, a cytoplasmic component of the flagellar type III protein export apparatus for efficient flagellar protein export and assembly [36,[39], [40], [41], [42]]. Val-111, Val-112 and Val-113 of FliN are responsible for the interaction with FliH (Fig. 2D) [39,41].
2.4. Subunit Organization in the C Ring Structure
Electron cryomicroscopy (cryoEM) image analysis has shown that the C ring structures of the purified CCW and CW motors have rotational symmetry varying from 32-fold to 35-fold, and the diameter varies accordingly [43,44]. The C ring diameters of the CCW and CW motors with C34 symmetry are 416 Å and 407 Å, respectively, and so the unit repeat distance along the circumference of the C ring is closer in the CW motor than in the CCW motor [45]. The C ring produced by a Salmonella fliF–fliG deletion fusion strain missing FliFC and FliGN lacks the inner lobe, suggesting that FliFC and FliGN together form the inner lobe (Fig. 1) [45,46]. In agreement with this, cryoEM images of the C ring containing the N-terminally green fluorescent protein (GFP) tagged FliG protein show an extra density corresponding to the GFP probe near the inner lobe [47]. The fliF–fliG deletion fusion results in unusual switching behavior of the flagellar motor, suggesting that the inner lobe is required for efficient and robust switching in the direction of flagellar motor rotation in response to changes in the environment [45]. The upper part of the C ring wall is formed by FliGM and FliGC. FliGM binds to FliGCN of its adjacent FliG subunit to produce a domain-swap polymer of FliG to form a ring in both CCW and CW motors [26,27,29]. Since HelixTorque of FliGCC interacts with MotAC [8,10], FliGCC is located at the top of the C ring wall (Fig. 1). Since FliMM directly binds to FliGM (Fig. 2C) [[18], [19], [20]], the continuous wall of the C ring with a thickness of 4.0 nm and a height of 6.0 nm is formed by side-by-side associations of the FliMM domains (Fig. 1) [32]. A continuous spiral density with a diameter of 7.0 nm along the circumference at the bottom edge of the C ring is made of FliMC and FliN (Fig. 1) [17,36].
3. Structural Basis for the Rotational Switching Mechanism
In E. coli and Salmonella, the flagellar motor is placed in a default CCW state [3,5]. Mutations located in and around HelixMC of FliG, which connects FliGM and FliGCN, cause unusual switching behavior of the flagellar motor [48], suggesting that helixMC is involved in switching the direction of flagellar motor rotation. HelixMC is located at the FliGM–FliMM interface and contributes to hydrophobic interactions between FliGM and FliMM (Fig. 3A) [18,19]. In-frame deletion of three residues, Pro-Ala-Ala at positions 169 to 171 of Salmonella FliG, which are located in HelixMC, locks the motor in the CW state even in the absence of CheY-P (CW-locked deletion) [49,50]. The crystal structure of the FliGM and FliGC domains derived from Thermotaoga maritima (Tm-FliGMC) with this CW-locked deletion have shown that the conformation of HelixMC is distinct from that of the wild-type [19,50,51]. In the wild-type Tm-FliGMC/Tm-FliMM complex, Val-172 of HelixMC of Tm-FliGMC makes hydrophobic contact with Ile-130 and Met-131 of Tm-FliMM (Fig. 3A) [18,19]. In contrast, disulfate crosslinking experiments have shown that HelixMC is dissociated from Tm-FliGM in the presence of the CW-locked deletion (Fig. 3A) [28]. Consistently, the CW-locked deletion of Tm-FliG reduces the binding affinity of Tm-FliGMC for Tm-FliMM by about 400-fold [28]. Therefore, it seems likely that the binding of CheY-P to FliM and FliN induces conformational rearrangements of the FliGM–FliMM interface, thereby causing dissociation of HelixMC from the interface to facilitate the remodeling of the FliG ring structure responsible for directional switching of the flagellar motor.
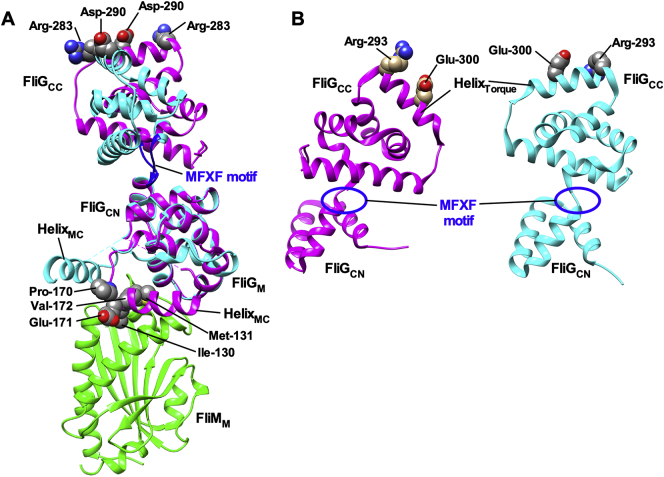
Fig. 3.
Structural basis for the switching mechanism. (A) Structural comparisons between wild-type FliGM and FliGC domains of T. maritima (Tm-FliGMC) and its CW-locked deletion variant, Tm-FliGMC(∆PEV). Cα ribbon drawing of Tm-FliGMC (magenta), Tm-FliGMC(∆PEV) (cyan) and Tm-FliMM (green). The FliGM domain of Tm-FliGMC(∆PEV) (PDB ID: 3AJC) was superimposed onto that of the Tm-FliGMC/Tm-FliMM complex (PDB ID: 4FHR). HelixMC is located at an interface between FliGM and FliMM. In contrast, the CW-locked deletion not only induces a distinct orientation of HelixMC relative to the FliGM–FliMM interface but also goes through a 90° rotation of FliGCC through a conserved MFXF motif colored in blue. Arg-283 and Asp-290 of Tm-FliG correspond to Arg-281 and Asp-289 of E. coli FliG, respectively. (B) Comparisons between the 3USY (cyan) and 3USW (magenta) structures of Helicobacter pylori FliG. Conformational rearrangements of the conserved MFXF motif induces a 180° rotation of FliGCC relative to FliGCN to reorient Arg-293 and Glu-300 residues, which correspond to Arg-281 and Asp-289 of E. coli FliG, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
HelixMC interacts with HelixNM connecting FliGN and FliGM (Fig. 2A) [23]. The E95D, D96V/Y, T103S, G106A/C and E108K substitutions in HelixNM of Salmonella FliG result in a strong CW switch bias [52]. A homology model of Salmonella FliG built based on the crystal structure of FliG derived from A. aeolicus (PDB code: 3HJL) has suggested that Thr-103 of HelixNM may make hydrophobic contacts with Pro-169 and Ala-173 of HelixMC [45]. These observations lead to a plausible hypothesis that a change in the HelixNM–HelixMC interaction mode may be required for conformational rearrangements of the C ring responsible for directional switching of the flagellar motor. A FliF–FliG full length fusion results in a strong CW switch bias of the E. coli flagellar motor [27]. Intragenic suppressor mutations, which improve the chemotactic behavior of the E. coli fliF–fliG full-length fusion strain, are located at the FliGN–FliGN interface [27], suggesting that a change in inter-molecular FliGN–FliGN interactions may be required for flagellar motor switching. Therefore, there is the possibility that conformational rearrangements of the FliGM–FliMM interface caused by the binding of CheY-P to the C ring influence the HelixNM–HelixMC interaction, thereby inducing conformational rearrangements of FliGN domains responsible for the switching in the direction of flagellar motor rotation.
The elementary process of torque generation by stator-rotor interactions is symmetric in CCW and CW rotation [12]. A hinge connecting FliGCN and FliGCC has a highly flexible nature at the conserved MFXF motif, allowing FliGCC to rotate 180° relative to FliGCN to reorient Arg-281 and Asp-289 residues in HelixTorque to achieve a symmetric elementary process of torque generation in both CCW and CW rotation (Fig. 3B) [[53], [54], [55], [56]]. Structural comparisons between Tm-FliGMC of the wild-type and Tm-FliGMC with the CW-locked deletion have shown that the CW-locked deletion induces a 90° rotation of FliGCC relative to FliGCN through the MFXF motif (Fig. 3A) [50]. Consistently, the binding of CheY-P to the C ring induces a tilting movement of FliMM, resulting in the rotation of FliGCC relative to FliGCN [34]. Therefore, it is possible that such a tilting movement of FliMM may promote a detachment of HelixMC from the FliGM–FliMM interface, resulting in the 180° rotation of FliGCC relative to FliGCN.
4. Adaptive remodeling of the C ring
FliM and FliN alternate their forms between localized and freely diffusing ones (Fig. 4), and the copy number of FliM and FliN in the CCW motor has been found to be about 1.3 times larger than that in the CW motor [[57], [58], [59], [60]]. Consistently, fluorescence anisotropy techniques have shown that the CCW motor accommodate more FliM1/FliN3 complexes without changing the spacing between FliM subunits [61]. Such exchanges depend on the direction of flagellar rotation but not on the binding of CheY-P to the C ring per se [58]. The timescale of this adaptive switch remodeling of the C ring structure is much slower (~ 1 min) than that of the rotational switching between the CCW and CW states (less than millisecond). Such a structural remodeling of the C ring is important for fine-tuning the chemotactic response to temporal changes in the environments [[62], [63], [64], [65]]. The CW-locked deletion of FliG considerably reduces the binding affinity of FliGM for FliMM presumably due to detachment of HelixMC from the FliGM–FliMM interface (Fig. 3A) [28]. Because FliM binds to HelixMC of FliG in the E. coli CCW motor [27], the dissociation of HelixMC from the FliGM–FliMM interface may promote the dissociation of several weakly bound FliM1/FliN3 complexes from the FliG ring when CheY-P binds to the C ring to switch from its CCW to CW states (Fig. 4).
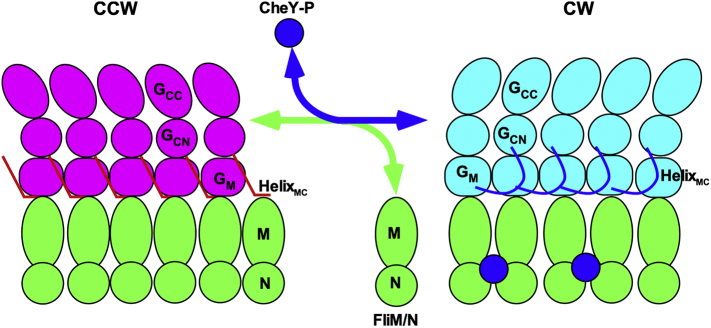
Fig. 4.
Adaptive remodeling of the FliG ring in the CCW and CW motors. Inter-molecular interactions of FliGCN with FliGM of its neighboring subunit produce the CCW ring structure. Upon binding of CheY-P to the C ring, conformational rearrangements of the FliGM–FliGC interface occur, resulting in detachment of HelixMC from the interface. As a result, several weakly bound FliM1/FliN3 complexes dissociate from the FliG ring.
5. Summary and Perspectives
Switching between the CW and CCW states of the flagellar motor is highly cooperative [66]. The cooperative switching mechanism can be explained by a conformational spread model, in which a switching event is mediated by conformational changes in a ring of subunits that spread from subunit to subunit via their interactions along the ring [[67], [68], [69]]. The binding of CheY-P to FliM and FliN affects subunit-subunit interactions between FliMM domains and between FliMC and FliN in the C ring to induce a 180° rotation of FliGCC relative to MotAC, thereby allowing the motor to rotate in CW direction [34]. HelixMC of FliG located at an interface between FliGM and FliMM plays an important role in highly cooperative remodeling of the FliG ring structure [28]. However, it remains unknown how HelixMC coordinates cooperative rearrangements of FliG subunits with changes in the direction of flagellar motor rotation. The C ring of the CCW motor can accommodate more FliM/FliN3 complexes without changing inter-subunit spacing, and directional switching of the motor induces several weakly bound FliM/FliN3 complexes from the C ring [[57], [58], [59], [60]]. Consistently, the CW-locked deletion weakens an interaction between FliGM and FliMM [28]. Because there is no difference in the rotational symmetry of the C ring between the purified CCW and CW motors [45], it remains unclear how several FliM1/FliN3 complexes weakly associate with the C ring when the motor spins in the CCW direction.
The elementary process of the torque-generation cycle is symmetrical in CCW and CW directions [12]. However, the output characteristics of the CW motor are distinct from those of the CCW motor. Torque produced by the CCW motor remains almost constant in a high-load, low-load regime of the torque-speed curve and decreases sharply to zero in a low-load, high-speed regime. In contrast, torque produced by the CW motor linearly decreases with increasing motor speed [70]. This suggests that directional switching of the flagellar motor may affect stator–rotor interactions in a load-dependent manner. However, nothing is known about the molecular mechanism. Furthermore, the switching rate of the flagellar motor also depends on the motor speed [71,72]. A recent non-equilibrium model of the flagellar motor switching has predicted that the motor sensitivity to CheY-P increases with an increase in motor torque [73]. However, it remains unknown how stator–rotor interactions modulate the binding affinity for CheY-P. High-resolution structural analysis of the C rings in the CCW and CW states by cryoEM image analysis will be essential to advance our mechanistic understanding of the directional switching mechanism of the flagellar motor.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgements
Our research is supported in part by the Japan Society for the Promotion of Science (JSPS KAKENHI Grant Numbers JP19H03182 to T.M., JP18K14638 to M.K. and JP25000013 to K.N.).
References
- 1.Berg H.C. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Morimoto Y.V., Minamino T. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules. 2014;4:217–234. doi: 10.3390/biom4010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minamino T., Imada K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015;23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Minamino T., Terahara N., Kojima S., Namba K. Autonomous control mechanism of stator assembly in the bacterial flagellar motor in response to changes in the environment. Mol Microbiol. 2018;109:723–734. doi: 10.1111/mmi.14092. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura S., Minamino T. Flagella-driven motility of bacteria. Biomoluecles. 2019;9:279. doi: 10.3390/biom9070279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J., Sharp L.L., Tang H.L., Lloyd S.A., Billings S. Function of protonatable residues in the flagellar motor of Escherichia coli: critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Che Y.S., Nakamura S., Morimoto Y.V., Kami-ike N., Namba K. Load-sensitive coupling of proton translocation and torque generation in the bacterial flagellar motor. Mol Microbiol. 2014;91:175–184. doi: 10.1111/mmi.12453. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J., Lloyd S.A., Blair D.F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci U S A. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morimoto Y.V., Nakamura S., Kami-ike N., Namba K., Minamino T. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol Microbiol. 2010;78:1117–1129. doi: 10.1111/j.1365-2958.2010.07391.x. [DOI] [PubMed] [Google Scholar]
- 10.Morimoto Y.V., Nakamura S., Hiraoka K.D., Namba K., Minamino T. Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J Bacteriol. 2013;195:474–481. doi: 10.1128/JB.01971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd S.A., Whitby F.G., Blair D.F., Hill C.P. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura S., Kami-ike N., Yokota J.P., Minamino T., Namba K. Evidence for symmetry in the elementary process of bidirectional torque generation by the bacterial flagellar motor. Proc Natl Acad Sci U S A. 2010;107:17616–17620. doi: 10.1073/pnas.1007448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueno T., Oosawa K., Aizawa S.I. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J Mol Biol. 1992;227:672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 14.Francis N.R., Sosinsky G.E., Thomas D., DeRosier D.J. Isolation, characterization, and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 15.Morimoto Y.V., Ito M., Hiraoka K.D., Che Y.S., Bai F., Kami-Ike N. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol Microbiol. 2014;91:1214–1226. doi: 10.1111/mmi.12529. [DOI] [PubMed] [Google Scholar]
- 16.Levenson R., Zhou H., Dahlquist F.W. Structural insights into the interaction between the bacterial flagellar motor proteins FliF and FliG. Biochemistry. 2012;51:5052–5060. doi: 10.1021/bi3004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDowell M.A., Marcoux J., McVicker G., Johnson S., Fong Y.H. Characterisation of Shigella Spa33 and Thermotoga FliM/N reveals a new model for C-ring assembly in T3SS. Mol Microbiol. 2016;99:749–766. doi: 10.1111/mmi.13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul K., Gonzalez-Bonet G., Bilwes A.M., Crane B.R., Blair D. Architecture of the flagellar rotor. EMBO J. 2011;30:2962–2971. doi: 10.1038/emboj.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vartanian A.S., Paz A., Fortgang E.A., Abramson J., Dahlquist F.W. Structure of flagellar motor proteins in complex allows for insights into motor structure and switching. J Biol Chem. 2012;287:35779–35783. doi: 10.1074/jbc.C112.378380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam K.H., Lam W.W., Wong J.Y., Chan L.C., Kotaka M., Ling T.K. Structural basis of FliG-FliM interaction in Helicobacter pylori. Mol Microbiol. 2013;88:798–812. doi: 10.1111/mmi.12222. [DOI] [PubMed] [Google Scholar]
- 21.Minamino T., Imada K., Namba K. Molecular motors of the bacterial flagella. Curr Opin Struct Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Stock D., Namba K., Lee L.K. Nanorotors and self-assembling macromolecular machines: the torque ring of the bacterial flagellar motor. Curr Opin Biotechnol. 2012;23:545–554. doi: 10.1016/j.copbio.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Lee K.L., Ginsburg M.A., Crovace C., Donohoe M., Stock D. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature. 2010;466:996–1000. doi: 10.1038/nature09300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch M.J., Levenson R., Kim E.A., Sircar R., Blair D.F., Dahlquist F.W. Co-folding of a FliF-FliG split domain forms the basis of the MS:C ring interface within the bacterial flagellar motor. Structure. 2017;25:317–328. doi: 10.1016/j.str.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue C., Lam K.H., Zhang H., Sun K., Lee S.H. Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly the motor MS–C ring in the bacterial flagellum. J Biol Chem. 2018;293:2066–2078. doi: 10.1074/jbc.M117.797936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker M.A., Hynson R.M., Ganuelas L.A., Mohammadi N.S., Liew C.W., Rey A.A. Domain-swap polymerization drives the self-assembly of the bacterial flagellar motor. Nat Struct Mol Biol. 2016;23:197–203. doi: 10.1038/nsmb.3172. [DOI] [PubMed] [Google Scholar]
- 27.Kim E.A., Panushka J., Meyer T., Carlisle R., Baker S., Ide N. Architecture of the flagellar switch complex of Escherichia coli: conformational plasticity of FliG and implications for adaptive remodeling. J Mol Biol. 2017;429:1305–1320. doi: 10.1016/j.jmb.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita M., Furukawa Y., Uchiyama S., Imada K., Namba K., Minamino T. Insight into adaptive remodeling of the rotor ring complex of the bacterial flagellar motor. Biochem Biophys Res Commun. 2018;496:12–17. doi: 10.1016/j.bbrc.2017.12.118. [DOI] [PubMed] [Google Scholar]
- 29.Kinoshita M., Namba K., Minamino T. Effect of a clockwise-locked deletion in FliG on the FliG ring structure of the bacterial flagellar motor. Genes Cells. 2018;23:241–247. doi: 10.1111/gtc.12565. [DOI] [PubMed] [Google Scholar]
- 30.Brown P.N., Terrazas M., Paul K., Blair D.F. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J Bacteriol. 2007;189:305–312. doi: 10.1128/JB.01281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathews M.A., Tang H.L., Blair D.F. Domain analysis of the FliM protein of Escherichia coli. J Bacteriol. 1998;180:5580–5590. doi: 10.1128/jb.180.21.5580-5590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S.Y., Lowder B., Bilwes A.M., Blair D.F., Crane B.R. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc Natl Acad Sci U S A. 2006;103:11886–11891. doi: 10.1073/pnas.0602811103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S.Y., Cho H.S., Pelton J.G., Yan D., Henderson R.K. Crystal structure of an activated response regulator bound to its target. Nat Struct Mol Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- 34.Paul K., Brunstetter D., Titen S., Blair D.F. A molecular mechanism of direction switching in the flagellar rotation of Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:17171–17176. doi: 10.1073/pnas.1110111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarkar M.K., Paul K., Blair D. Subunit organization and reversal associated movements in the flagellar switch of Escherichia coli. J Biol Chem. 2010;285:675–684. doi: 10.1074/jbc.M109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Notti R.Q., Bhattacharya S., Lilic M., Stebbins C.E. A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat Commun. 2015;6:7125. doi: 10.1038/ncomms8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown P.N., Mathews M.A., Joss L.A., Hill C.P., Blair D.F. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol. 2005;187:2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar M.K., Paul K., Blair D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:9370–9375. doi: 10.1073/pnas.1000935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul K., Harmon J.G., Blair D.F. Mutational analysis of the flagellar rotor protein FliN: identification of surfaces important for flagellar assembly and switching. J Bacteriol. 2006;188:5240–5248. doi: 10.1128/JB.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González-Pedrajo B., Minamino T., Kihara M., Namba K. Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol Microbiol. 2006;60:984–998. doi: 10.1111/j.1365-2958.2006.05149.x. [DOI] [PubMed] [Google Scholar]
- 41.McMurry J.L., Murphy J.W., González-Pedrajo B. The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry. 2006;45:11790–11798. doi: 10.1021/bi0605890. [DOI] [PubMed] [Google Scholar]
- 42.Minamino T., Yoshimura S.D.J., Morimoto Y.V., González-Pedrajo B., Kami-ike N. Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol Microbiol. 2009;74:1471–1483. doi: 10.1111/j.1365-2958.2009.06946.x. [DOI] [PubMed] [Google Scholar]
- 43.Thomas D.R., Morgan D.G., DeRosier D.J. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci U S A. 1999;96:10134–10139. doi: 10.1073/pnas.96.18.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas D.R., Francis N.R., Xu C., DeRosier D.J. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakai T., Miyata T., Terahara N., Mori K., Inoiue Y. Novel insights into conformational rearrangements of the bacterial flagellar switch complex. mBio. 2019;10 doi: 10.1128/mBio.00079-19. e00079–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas D., Morgan D.G., DeRosier D.J. Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J Bacteriol. 2001;183:6404–6412. doi: 10.1128/JB.183.21.6404-6412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morimoto Y.V., Kami-ike N., Miyata T., Kawamoto A., Kato T. High-resolution pH imaging of living bacterial cell to detect local pH differences. mBio. 2016;7:01911–01916. doi: 10.1128/mBio.01911-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Way S.M., Millas S.G., Lee A.H., Manson M.D. Rusty, jammed, and well-oiled hinges: mutations affecting the interdomain region of FliG, a rotor element of the Escherichia coli flagellar motor. J Bacteriol. 2006;188:3944–3951. doi: 10.1128/JB.186.10.3173-3181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Togashi F., Yamaguchi S., Kihara M., Aizawa S.I., Macnab R.M. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J Bacteriol. 1997;179:2994–3003. doi: 10.1128/jb.179.9.2994-3003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minamino T., Imada K., Kinoshita M., Nakamura S., Morimoto Y.V., Namba K. Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown P.N., Hill C.P., Blair D.F. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 2002;21:3225–3234. doi: 10.1093/emboj/cdf332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irikura V.M., Kihara M., Yamaguchi S., Sockett H., Macnab R.M. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol. 1993;175:802–810. doi: 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam K.H., Ip W.S., Lam Y.W., Chan S.O., Ling T.K., Au S.W. Multiple conformations of the FliG C-terminal domain provide insight into flagellar motor switching. Structure. 2012;20:315–325. doi: 10.1016/j.str.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 54.Pandini A., Morcos F., Khan S. The gearbox of the bacterial flagellar motor switch. Structure. 2016;24:1209–1220. doi: 10.1016/j.str.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyanoiri Y., Hijikata A., Nishino Y., Gohara M., Onoue Y., Kojima M. Structural and functional analysis of the C-terminal region of FliG, an essential motor component of Vibrio Na+-driven flagella. Structure. 2017;25:1540–1548. doi: 10.1016/j.str.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Nishikino T., Hijikata A., Miyanoiri Y., Onoue Y., Kojima S., Shirai T. Rotational direction of flagellar motor from the conformation of FliG middle domain in marine Vibrio. Sci Rep. 2018;8 doi: 10.1038/s41598-018-35902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delalez N.J., Wadhams G.H., Rosser G., Xue Q., Brown M.T., Dobbie I.M. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc Natl Acad Sci U S A. 2010;107:11347–11351. doi: 10.1073/pnas.1000284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lele P.P., Branch R.W., Nathan V.S., Berg H.C. Mechanism for adaptive remodeling of the bacterial flagellar switch. Proc Natl Acad Sci U S A. 2012;109:20018–20022. doi: 10.1073/pnas.1212327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delalez NJ, Berry RM, Armitage JP. Stoichiometry and turnover of the bacterial flagellar switch protein FliN. mBio 2014;5:e01216–14. [DOI] [PMC free article] [PubMed]
- 60.Branch R.W., Sayegh M.N., Shen C., Nathan V.S., Berg H.C. Adaptive remodeling by FliN in the bacterial rotary motor. J Mol Biol. 2014;426:3314–3324. doi: 10.1016/j.jmb.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hosu B.G., Berg H.C. CW and CCW conformations of the E. coli flagellar motor C-ring evaluated by fluorescence anisotropy. Biophys J. 2018;114:641–649. doi: 10.1016/j.bpj.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan J., Branch R.W., Hosu B.G., Berg H.C. Adaptation at the output of the chemotaxis signalling pathway. Nature. 2012;484:233–236. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan J., Berg H.C. Ultrasensitivity of an adaptive bacterial motor. J Mol Biol. 2013;425:1760–1764. doi: 10.1016/j.jmb.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dufour Y.S., Fu X., Hernandez-Nunez L., Emonet T. Limits of feedback control in bacterial chemotaxis. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lele P.P., Shrivastava A., Roland T., Berg H.C. Response thresholds in bacterial chemotaxis. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cluzel P., Surette M., Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 67.Duke T.A., Le Novere N., Bray D. Conformational spread in a ring of proteins: a stochastic approach to allostery. J Mol Biol. 2001;308:541–553. doi: 10.1006/jmbi.2001.4610. [DOI] [PubMed] [Google Scholar]
- 68.Bai F., Branch R.W., Nicolau D.V., Jr., Pilizota T., Steel B.C., Maini P.K. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science. 2010;327:685–689. doi: 10.1126/science.1182105. [DOI] [PubMed] [Google Scholar]
- 69.Bai F., Minamino T., Wu Z., Namba K., Xing J. Coupling between switching regulation and torque generation in bacterial flagellar motor. Phys Rev Lett. 2012;108 doi: 10.1103/PhysRevLett.108.178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan J., Farner K.A., Turner L., Berg H.C. Asymmetry in the clockwise and counterclockwise rotation of the bacterial flagellar motor. Proc Natl Acad Sci U S A. 2010;107:12846–12849. doi: 10.1073/pnas.1007333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fahrner K.A., Ryu W.S., Biomechanics Berg H.C. Bacterial flagellar switching under load. Nature. 2003;423:938. doi: 10.1038/423938a. [DOI] [PubMed] [Google Scholar]
- 72.Yuan J., Fahrner K.A., Berg H.C. Switching of the bacterial flagellar motor near zero load. J Mol Biol. 2009;390:394–400. doi: 10.1016/j.jmb.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang F., Shi H., He R., Wang R., Zhang R., Yuan J. Non-equilibrium effect in the allosteric regulation of the bacterial flagellar switch. Nat Phys. 2017;13:710–714. [Google Scholar]