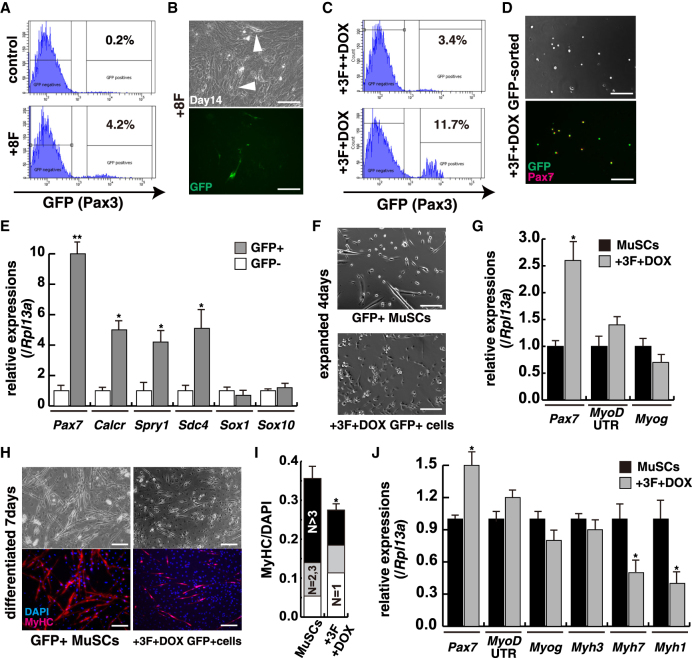
Figure 2.
The Combination of Four Transcription Factors Induces PAX3-Expressing Myogenic Cells from Mouse Fibroblasts
(A and B) Detectable GFP expression in MEFs (arrowheads) infected with eight transcription factors (+8F) after 14 days.
(C) FACS analyses of GFP-expressing cells with mouse Pax3, HeyL, and Klf4 (+3F) and persistent MYOD (++DOX) or transient MYOD for 72 h (+DOX).
(D) Immunofluorescence for GFP (labeled with Alexa 488, green) and Pax7 (labeled with Alexa 647, red) with GFP-positive cells induced by 3F and transient MYOD acceleration (+3F+DOX).
(E) Expression levels of Pax7, Calcr, Spry1, Sdc4, Sox1, and Sox10 transcripts levels in induced Pax3-GFP-positive (gray) or -negative (white) cells with 3F+DOX. n = 3 independent replicates; p values are determined by t test from a two-tailed distribution. ∗∗p < 0.01, ∗p < 0.05.
(F) Morphological features of cultured GFP-positive satellite cells (GFP+ MuSCs; upper panel) and three transcription factors with transient DOX treatment (+3F+DOX GFP+ cells; lower panel) for 4 days.
(G) Myogenic transcripts of Pax7, endogenous MyoD (MyoD UTR), and Myog relative to Rpl13a transcripts in GFP+ cells cultured for 4 days. n = 3 independent replicates; p values are determined by t test from a two-tailed distribution. ∗p < 0.05.
(H) Differentiated myogenic cells (DAPI, blue; MyHC, Alexa 647, red) from induced GFP-positive cells with 3F+DOX (right panels) compared with mouse Pax3-GFP satellite cells (GFP+ MuSCs; left panels) on 2% horse serum for 7 days.
(I) Quantification of the ratio of DAPI-positive mono (N = 1) or multiple nuclei (N = 2, 3 or N > 3) present in single MyHC-positive myofibers of (H). n = 3 independent replicates; p values are determined by t test from a two-tailed distribution. ∗p < 0.05.
(J) Transcriptional levels of myogenic markers Pax7, MyoD, Myog, Myh3, Myh7, and Myh1, differentiated for 7 days from GFP+ cells. n = 3 independent replicates; p values are determined by t test from a two-tailed distribution. ∗p < 0.05.
Error bars indicate ±SEM. Scale bars, 50 μm.