Figure 1.
Introduction of iPSC Lines Used in This Study
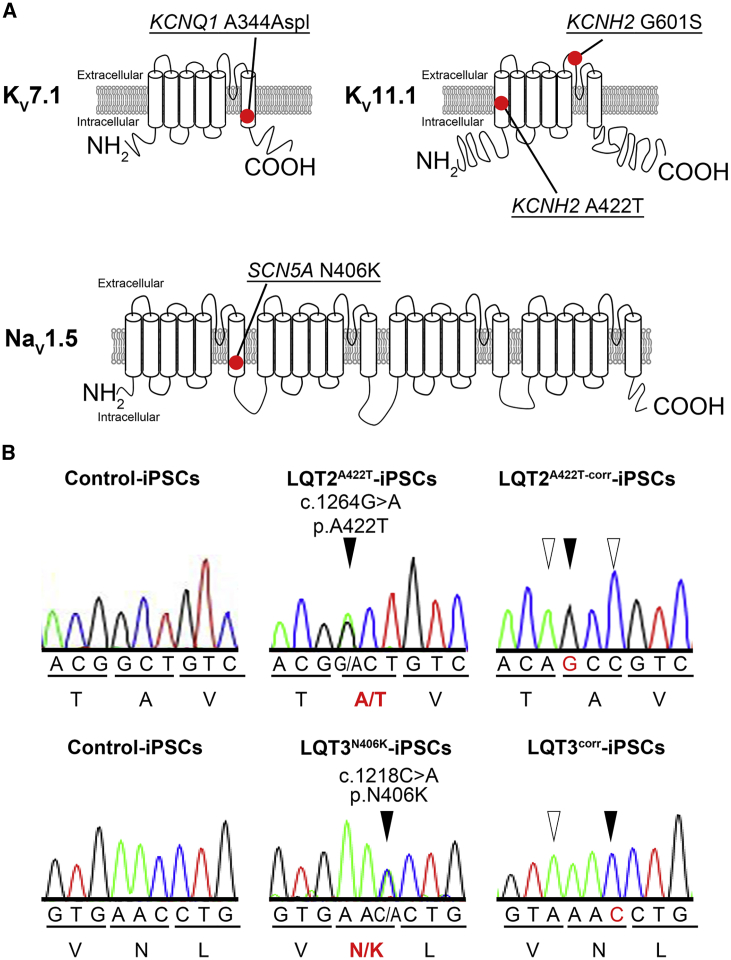
(A) Summary of topologies of mutations of iPSC lines used in this study. LQT1A344Aspl carries a heterozygous KCNQ1 mutation (c.1032C > A, p.A344Aspl); LQT2A422T and LQT2A422T-corr are the isogenic pair harboring the heterozygous KCNH2 mutation (c.1264G > A, p.A422T) and the corrected sequence, respectively; LQT2G601S carries a heterozygous KCNH2 mutation (c.1801G > A, p.G601S); and LQT3N406K and LQT3corr are the isogenic pair harboring the heterozygous SCN5A mutation (c.1218C > A, p.N406K) and the corrected sequence, respectively.
(B) Sequence analysis of PCR-amplified genomic DNA of the isogenic pair of LQT2A422T and LQT2 A422T-corr and of LQT3N406K and LQT3corr, respectively. The gene-corrected cell lines harbor several silent mutations (white arrow head) to avoid further digestion by CRISPR/Cas9.