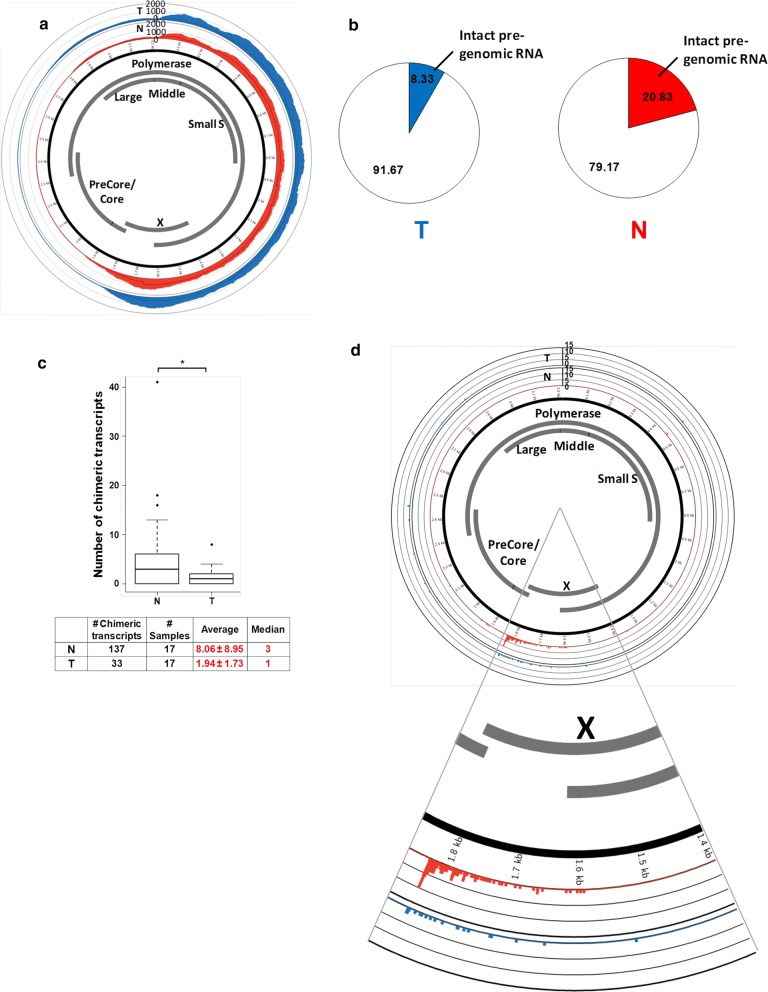
Fig. 3.
Coverage of HBV genome and proportion of patients with potentially intact pre-genomic RNA. a Coverage of HBV genome by sequencing reads. The circos plot shows the average coverage of each nucleotide of HBV genome by the sequencing reads. Red and blue histograms show the average coverage in the 24 N and T, respectively. Coverage is significantly higher in the Pre-S and X genes. b Proportion of patients with potentially intact pre-genomic RNA. As the pre-genomic RNA is 3.5 kb and covers the entire HBV genome, the pre-genomic RNA is considered incomplete if any region of the HBV is not detected. Intact pre-genomic RNA is likely to be found in 20.83% and 8.33% of N and T samples, respectively, suggesting that HBV replication is not a common event in HCC patient liver. c Less variety of chimeric transcripts in the tumor of HCC patients. Boxplot showing average and median number of different chimeric transcripts. The table shows the total number of different chimeric transcripts, number of samples which contain chimeric transcripts, the average and median number of chimeric transcripts detected in T and N samples. The number of different chimeric transcripts detected in T is significantly lower in compared to the N samples (p-value < 0.05, paired t-test) suggesting that a subset of functional chimeric transcripts is selected in the process of tumorigenesis. d Circos plot showing the distribution of fusion sites on HBV genome. The fusion sites between the HBV and host sequences in the chimeric transcripts are significantly located in the region between 1600 and 1900 of the HBV genome (near the end of the HBx gene) in both non-tumor and tumor samples (p-value < 0.001, random sampling test)