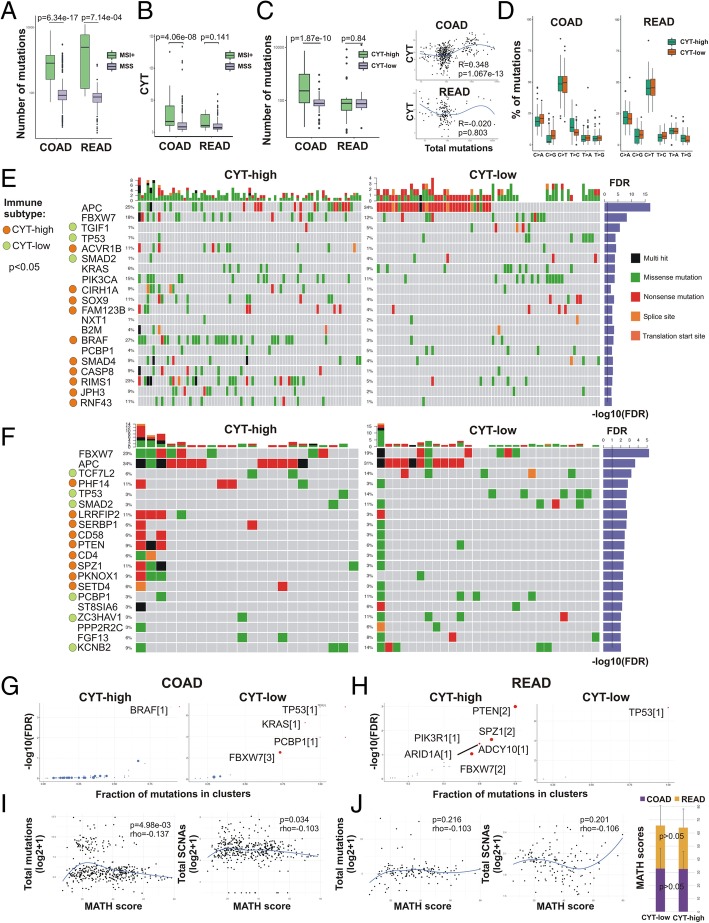
Fig. 3.
a The mutation load increased considerably in colorectal tumors with high microsatellite instability (MSI) vs stable microsatellites (MSS). b Cytolytic activity (CYT) is considerably higher in colon (but not rectal) tumors with high microsatellite instability (MSI+) vs MSS tumors. c The mutation load increased considerably in CYT-high colon (but not rectal) tumors and was significantly correlated with the cytolytic index (CYT). d Nonsynonymous mutation spectra across COAD and READ cytolytic subsets, depicting the percentage (%) of each mutation type in high and low colon and rectal tumors, respectively. Most mutations across the datasets were associated with C > T (and G > A) transitions and the frequency of specific substitutions did not differ between CYT-high and CYT-low tumors. e-f Co-mutation plot showing significantly mutated genes (SMGs, FDR < 0.1) in cytolytic subsets of colon (COAD) (e) and rectal (READ) tumors (f). Green, red, pink, black and orange boxes indicate missense, nonsense, transcription start site, multi-hit and splice-site mutations, respectively. SMGs that correlate with immune cytolytic subtypes (p < 0.05) are highlighted by green or orange circles in the left columns of each dataset. Each SMG’s q-values (−log10(FDR)) are plotted as a right-side bar plot in blue color. g-h Plots show the different cancer driver genes in the two cytolytic subgroups of COAD and READ adenocarcinomas, using OncodriveCLUST [42]. Cancer driver genes are depicted as scatter plots, in which the size of the points is proportional to the number of clusters found in the corresponding gene. The x-axis shows the fraction of mutations observed in these clusters. Scores within brackets next to the gene names denote the number of the gene’s mutational clusters. i-j Tumor heterogeneity and mutation load in CRC. The width of each tumor’s variant allele frequency (VAF) distribution (MATH scores) does not correlate with the total mutation count or the total copy number events, neither in colon (i) nor in rectal cancers (j). No difference in MATH scores between cytolytic subsets in COAD and READ cohorts (p > 0.05). P-values were calculated using Kruskal-Wallis test (j)