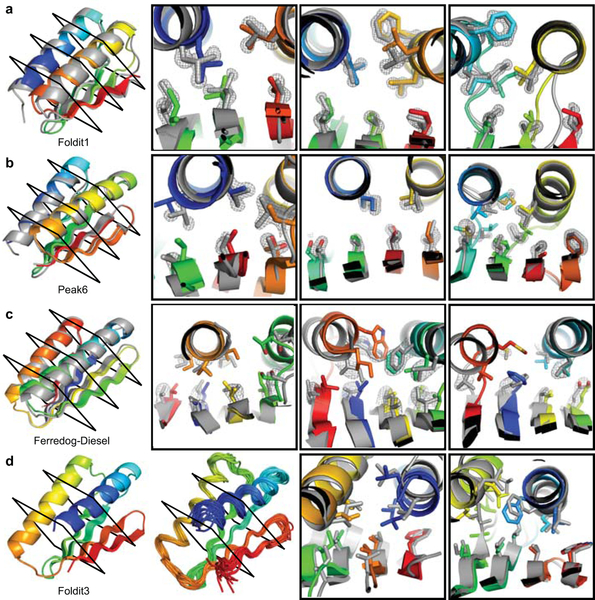
Figure 4. High-resolution structures of Foldit player-designed proteins.
a, The Foldit1 design (fold V in Fig 3: 3 β-strands with sheet order 1–2–3) model backbone (rainbow) aligns to the crystal structure (gray) with Cα-RMSD of 1.1 Å. b, The Peak6 design (fold III: 4 strands, sheet order 1-2-4-3) model backbone aligns to the crystal structure with Cα-RMSD of 0.9 Å. c, The Ferredog-Diesel design (fold I: 4 strands, sheet order 4-1-3-2) model backbone aligns to the crystal structure with Cα-RMSD of 1.7 Å. Cross-sections show core residue sidechains, with the composite omit 2mFo-DFc map contoured at 2.0 σ (a-b) or 1.0 σ (c). d, The Foldit3 design model (fold XVIII: 4 strands, sheet order 2-1-3-4) and NMR ensemble. The design model aligns to the representative (medoid) NMR model with a Cα-RMSD of 1.1 Å. Cross sections compare core side chains in the design model (rainbow) and representative NMR model (gray).