Abstract
Counterfeiting is an incredibly widespread problem, with some estimates placing its economic impact above 2% of worldwide GDP. The scale of the issue suggests that current preventive measures are either technologically insufficient or too impractical and costly to be widely adopted. High-density arrays of biomolecules are explored here as security devices that can be coupled to a valuable commodity as proof of its authenticity. Light-directed DNA array fabrication technology is used to synthesize arrays that are designed to resist analysis with sequencing-by-hybridization approaches. A relatively simple sequence design strategy forces a counterfeiter to undertake a prohibitively high number of complex experiments to decipher the array sequences employed.
Keywords: Encryption, Anti-counterfeiting, Authentication, Photolithography, Maskless array synthesis
Graphical Abstract
The storage and transmission of biological information is the fundamental role of genetic material. As DNA sequencing and synthesis costs have fallen, a variety of encoding schemes have been developed to repurpose DNA as a medium to store arbitrary forms of data. Many of these are concerned with increasing the information storage capacity of DNA, accurate retrieval of the information, or chemical means of preserving the integrity of the DNA molecules.1–4 A small, yet diverse, group of approaches have also sought to restrict access to the underlying data,5–10 though relatively few examples of such DNA cryptography have entered routine use. Here, a maskless array synthesizer is used for disguising information in a high-density DNA array in an analogy to key-based encryption. The strategy relies upon rendering the arrays resistant to sequencing-by-hybridization techniques which could otherwise be employed for decryption. The approach is explored as an anti-counterfeiting technology.
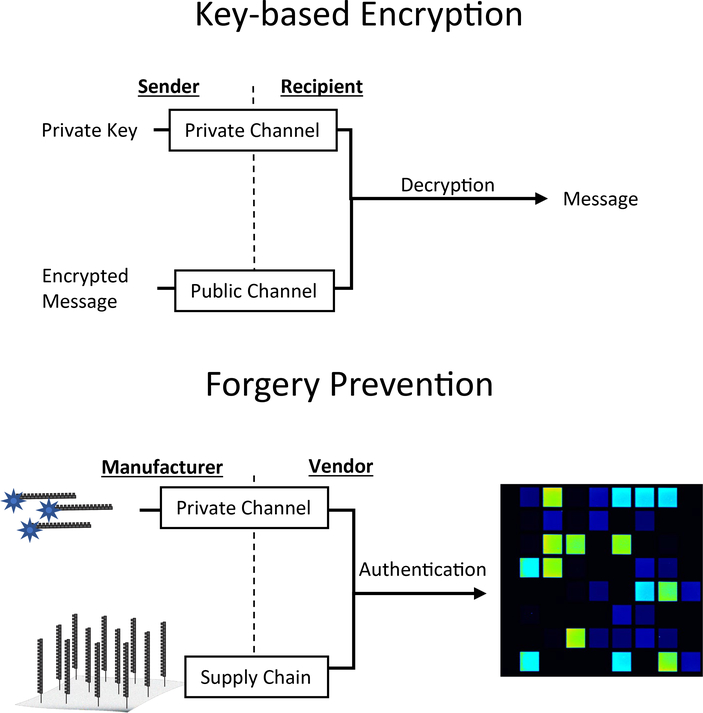
Scheme 1 outlines how an array-based system could be used to ensure the authenticity of a product. Under the assumption that there is a secure mode of communication, fluorescently-labeled oligonucleotides act as a private ‘key’ to reveal the information in the array upon hybridization. Each location on the surface exhibits a distinct signal, for multiple fluorescence channels, while the arrangement of the array features can form shapes such as barcodes, lot numbers, or watermarks. Despite this appeal, existing DNA-based anti-counterfeiting approaches have not utilized the array itself as the marker of authenticity.11, 12 This is likely because the cost of array fabrication has been too high for routine use with inexpensive goods, yet they are too vulnerable to forgery to secure high-value items. Improvements in millichip technology and increased array manufacture speeds suggest opportunities for drastic enhancements to array fabrication throughput, but there has not yet been an approach for rendering the arrays themselves resistant to analysis and forgery.13–17
Scheme 1.
Analogy of private key encryption to an array-based anti-forgery device.
In order for a DNA array to be replicated, a forger would need to determine both the identities of the sequences and their spatial locations on the surface. Sequencing by hybridization (SBH) is the most general route to decrypt an array in that it would allow a forger to decipher arrays even of unnatural nucleic acids, such as LNA, L-DNA, or PNA, that are not amenable to current sequencing-by-synthesis approaches.18–20 A drawback of SBH is that de novo sequencing by SBH requires hybridization with relatively short oligonucleotides (reads) so that complete coverage can be achieved in a tractable number of hybridization cycles. The short reads cause a loss of long-range sequence information that can produce ambiguities during assembly, particularly when applied to repetitive sequences. We make use of this limitation of SBH, and further complicate the task of a would-be forger by placing two related sequences within each array feature.
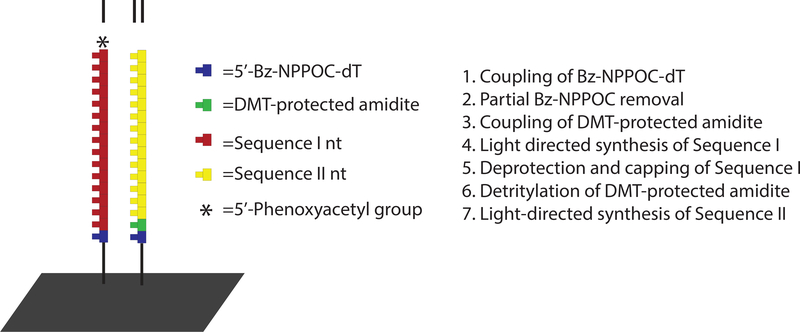
Arrays containing two sequences per feature can be produced using a maskless fabrication approach that has previously been applied to SNP detection and RNA array fabrication.14, 21, 22 Briefly, the extent of photodeprotection is controlled at an early step of the fabrication process so that an orthogonal protecting group is installed, typically by coupling a dimethoxytrityl-protected phosphoramidite, on approximately half of the molecules populating a given site. This protecting group remains attached throughout the light-directed synthesis of the first sequence and is then removed so the second sequence can be generated with the remaining fraction of site density. Scheme 2 provides more detail on this approach.
Scheme 2:
Synthesis approach for arrays containing two sequences per feature.
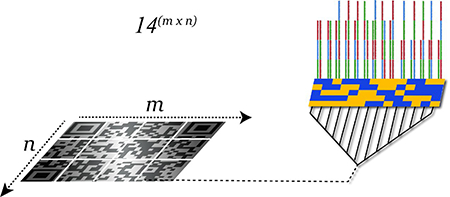
An overview of the sequence design is shown in Scheme 3. The concept is to use pairs of sequences at each site so that hybridizations with short oligonucleotides produce reads with multiple possible assemblies. This renders the elucidation of the surface-bound sequences using SBH much more difficult. Consider the parent string ABCAC where A, B, and C are blocks of sequence each of length l. Reads generated from iterative hybridizations to the ABCAC string can identify the sequences of the blocks as well as the junctions between adjacent blocks. If the read length, r, is less than l, a single read cannot span more than one junction simultaneously, so the overall string must be inferred from the block junctions (AB, BC, CA, and AC in the above example). If one employs pairs of sequences made from the same A,B,C sequence blocks in each feature, this design yields 14 configurations that 1) give the same hybridization signature as the parent string when r≤l (making decryption difficult for the would-be forger) and 2) can be uniquely identified by hybridization of sequences of length ≥3 × l (making authentication easy for those with the hybridization key). For an array comprised of N features, this design produces 14N possible assemblies for the SBH reads generated when r≤l.
Scheme 3:
Sequence design strategy. In the upper left, an example of A, B, and C sequence blocks with a length of 10 nt each, composed into the ABCAC string. The theoretical reads generated from sequencing by hybridization using pentamers are shown below the string. The table at the right provides the architecture (‘Block structure’) of pairs of sequences which yield the same theoretical SBH reads as the ABCAC string. The ‘Binding Pattern’ gives the design of fluorescently-labelled three-block sequences that when hybridized to the array will distinguish the 14 array features from one another. Subscript ‘C’ indicates complementarity to the sequence of the given block structure (‘ABCC’ is the complement of sequence ABC). Yellow denotes hybridization signal will be observed, blue indicates no hybridization signal will be observed.
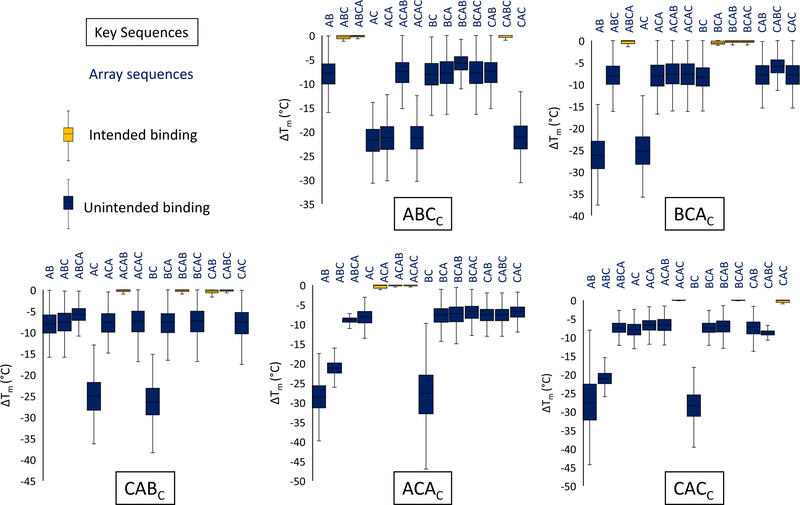
An underlying assumption is that, when there is little homology between A, B, and C, hybridization conditions exist which can differentiate between the binding of perfectly matched sequences and those which are ~2/3 complementary. The strategy was investigated computationally to assess the generality of the approach. The 14 feature types shown in Scheme 3 are comprised of 14 different surface oligonucleotides. A, B, and C blocks were randomly generated and used to create the private key sequences and surface sequences. The hybridization specificity was then assessed using the DINAMelt server’s two-state function with DNA energy parameters at 37° C, and 150 mM Na+ concentration.23 Each private key sequence was tested against the set of the 14 surface sequences for every assignment of A, B, and C. The Tm difference (ΔTm) between the highest calculated value and the other members of the set was then determined. Figure 1 shows the average ΔTm for 500 randomly generated sets of A, B, and C blocks 10 nt in size. All the average ΔTm values between the undesigned and designed interactions differed by more than 5° C, indicating a general basis to observe the expected differential hybridization. Approximately 70% of the assignments tested had no undesigned ΔTm values above −3° C for all key and surface block combinations.
Figure 1:
Calculated average Tm differences for hybridization of key oligonucleotides to their targeted surface complements versus other partially complementary background sequences. Box denotes the interquartile range and whiskers denote outermost values.
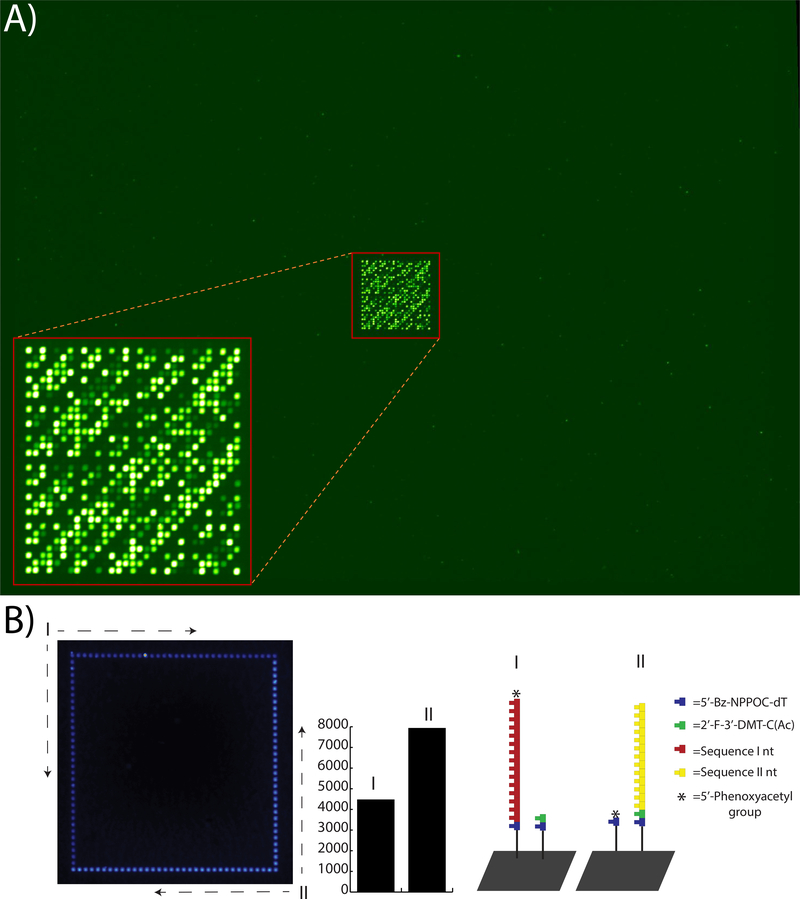
This approach is so powerful because the forger does not know the composition of the private key and must decipher every feature on an array to guarantee a successful replicate. Figure 2A shows the hybridization of an oligonucleotide key to an array comprised of 87,164 features while Figure 2B depicts the quality control elements used as a measure of the relative population of the sequences within the features. The key is comprised of six Hex-labelled oligonucleotides 32 nt in length and of the form ABCC. The foreground pattern shown in the inset of 2A was fashioned after the Hadamard matrix used to communicate with the Mariner 9 spacecraft and confirms the authenticity of the array.24 However, the would-be forger would not be able to determine the identities of the sequences that comprise the array, and thus would be unable to fabricate a replicate. The background sequences, where hybridization signal is not observed, were designed using randomly generated sequences of block sizes of either 10 or 11 nt, so that reads 10 bases or shorter produce data consistent with 1487164 or 1.27 × 1099901 possible array designs. SBH with 11 nt read lengths would require over 4 million hybridization events, rendering decryption impractical.
Figure 2.
A) Hybridization of a private key to an array comprised of over 87,000 features. In principle, the hybridization behavior of any combination of features throughout the array may act as an indicator of authenticity. B) Measurement of relative populations of 1st and 2nd strands. The inset shown in (A) was bordered by replicate features as shown on the left-hand side. Half of the edges were synthesized as part of the first set of sequences (‘I’), while the remainder were synthesized as part of the second set (‘II’). Hybridization with a 5’-Cy5 labelled oligonucleotide was used to determine that the proportion of sequence I to sequence II is approximately 1:2 for this example. The DMT-protected amidite used here was a 2’-F-3’-DMT-rC (Ac) 5’-phosphoramidite.
Developing a defense against SBH is a significant step towards adapting arrays as an anti-counterfeiting tool. Though it is not possible to rule out future techniques which could decrypt such arrays, they could still function as effective deterrents if the cost of decryption exceeds the value of a particular good. Even arrays with a few thousand sequences may be impractical to decrypt, suggesting a new application for millichip technology and related polymeric array substrates.13, 14, 25 The sequence design strategy described here is quite general, applies to any nucleic acid-based polymer, and could conceivably extend to other array fabrication platforms. Any oligonucleotide chemistry which produces a discontinuity in a long sequence, such as a long spacer or inversion of polarity, would likely achieve the same result as placing two sequences within a feature. This would allow SBH-resistant arrays to be made using DMT-protected phosphoramidites on inkjet platforms. Spotted array platforms may be especially useful as well because they can utilize oligonucleotides of a length beyond the limits of in situ chemistry. This would enable blocks of high l, more complicated block arrangement schemes using longer strings, or features comprised of three or more distinct sequences for enhanced levels of security.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants 1RO1GM108727, 1RO1GM109099, and 5T32GM08349.
Footnotes
Supporting Information.
The following files are available free of charge.
Supplemental data (quality control data and results of hybridization calculations), materials and methods (PDF)
Calculated melting temperature raw data (Excel)
Table of array sequences (Excel)
REFERENCES
- 1.Erlich Y; Zielinski D, DNA Fountain Enables a Robust and Efficient Storage Architecture. Science 2017, 355 (6328), 950–954. [DOI] [PubMed] [Google Scholar]
- 2.Grass RN; Heckel R; Puddu M; Paunescu D; Stark WJ, Robust Chemical Preservation of Digital Information on DNA in Silica with Error-Correcting Codes. 2015, 54 (8), 2552–2555. [DOI] [PubMed] [Google Scholar]
- 3.Church GM; Gao Y; Kosuri S, Next-Generation Digital Information Storage in DNA. Science 2012, 337 (6102), 1628. [DOI] [PubMed] [Google Scholar]
- 4.Goldman N; Bertone P; Chen S; Dessimoz C; LeProust EM; Sipos B; Birney E, Toward Practical High-Capacity Low-Maintenance Storage of Digital Information in Synthesised DNA. Nature 2013, 494 (7435), 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandrasekaran AR; Levchenko O; Patel DS; MacIsaac M; Halvorsen K, Addressable Configurations of DNA Nanostructures for Rewritable Memory. Nucleic Acids Research 2017, 11459–11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halvorsen K; Wong WP, Binary DNA Nanostructures for Data Encryption. PLOS ONE 2012, 7 (9), e44212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gehani A; LaBean T; Reif J, DNA-based Cryptography In Aspects of Molecular Computing Jonoska N; Păun G; Rozenberg G, Eds. Springer Berlin Heidelberg: Berlin, Heidelberg, 2004; pp 167–188. [Google Scholar]
- 8.Clelland CT; Risca V; Bancroft C, Hiding Messages in DNA microdots. Nature 1999, 399, 533. [DOI] [PubMed] [Google Scholar]
- 9.Shoshani S; Piran R; Arava Y; Keinan E, A Molecular Cryptosystem for Images by DNA Computing. Angewandte Chemie 2012, 51 (12), 2883–2887. [DOI] [PubMed] [Google Scholar]
- 10.Arppe R; Sørensen TJ, Physical Unclonable Functions Generated Through Chemical Methods for Anti-Counterfeiting. Nature Reviews Chemistry 2017, 1, 0031. [Google Scholar]
- 11.Butland CL; Baggot B Labeling Technique for Countering Product Diversion and Product Counterfeiting. US 6030657A, November 1, 1994 [Google Scholar]
- 12.Lawrence J; Liang B In-field DNA Extraction, Detection and Authentication Methods and Systems Therefor. CA 2959312A1, August 28, 2014. [Google Scholar]
- 13.Heinrich KW; Wolfer J; Hong D; LeBlanc M; Sussman MR, DNA Millichips as a Low-Cost Platform for Gene Expression Analysis. Plant Physiology 2012, 159 (2), 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holden MT; Carter MCD; Wu C-H; Wolfer J; Codner E; Sussman MR; Lynn DM; Smith LM, Photolithographic Synthesis of High-Density DNA and RNA Arrays on Flexible, Transparent, and Easily Subdivided Plastic Substrates. Analytical Chemistry 2015, 87 (22), 11420–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sack M; Hölz K; Holik A-K; Kretschy N; Somoza V; Stengele K-P; Somoza MM, Express Photolithographic DNA Microarray Synthesis with Optimized Chemistry and High-Efficiency Photolabile Groups. Journal of Nanobiotechnology 2016, 14 (1), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kretschy N; Holik A-K; Somoza V; Stengele K-P; Somoza MM, Next-Generation o-Nitrobenzyl Photolabile Groups for Light-Directed Chemistry and Microarray Synthesis. Angewandte Chemie 2015, 54 (29), 8555–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sack M; Kretschy N; Rohm B; Somoza V; Somoza MM, Simultaneous LightDirected Synthesis of Mirror-Image Microarrays in a Photochemical Reaction Cell with Flare Suppression. Analytical Chemistry 2013, 85 (18), 8513–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauser NC; Martinez R; Jacob A; Rupp S; Hoheisel JD; Matysiak S, Utilising the Left-Helical Conformation of L-DNA for Analysing Different Marker Types on a Single Universal Microarray Platform. Nucleic Acids Research 2006, 34 (18), 5101–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z-C; Shin D-S; Shokouhimehr M; Lee K-N; Yoo B-W; Kim Y-K; Lee Y-S, Light-Directed Synthesis of Peptide Nucleic Acids (PNAs) Chips. Biosensors and Bioelectronics 2007, 22 (12), 2891–2897. [DOI] [PubMed] [Google Scholar]
- 20.Yang F; Dong B; Nie K; Shi H; Wu Y; Wang H; Liu Z, Light-Directed Synthesis of High-Density Peptide Nucleic Acid Microarrays. ACS Combinatorial Science 2015, 17 (10), 608–614. [DOI] [PubMed] [Google Scholar]
- 21.Nie B; Yang M; Fu W; Liang Z, Surface Invasive Cleavage Assay on a Maskless Light-Directed Diamond DNA Microarray for Genome-Wide Human SNP Mapping. Analyst 2015, 140 (13), 4549–4557. [DOI] [PubMed] [Google Scholar]
- 22.Wu C-H; Holden MT; Smith LM, Enzymatic Fabrication of High Density RNA Arrays. Angewandte Chemie 2014, 53 (49), 13514–13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markham NR; Zuker M, DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Research 2005, 33 (Web Server issue), W577–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agaian SS; Sarukhanyan HG; Egiazarian KO; Astola J, Hadamard Transforms. SPIE Press: 2011; Vol. PM207, p 520.. [Google Scholar]
- 25.Holden MT; Carter MCD; Ting SK; Lynn DM; Smith LM, Parallel DNA Synthesis on Poly(ethylene terephthalate). ChemBioChem 2017, 18 (19), 1914–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.