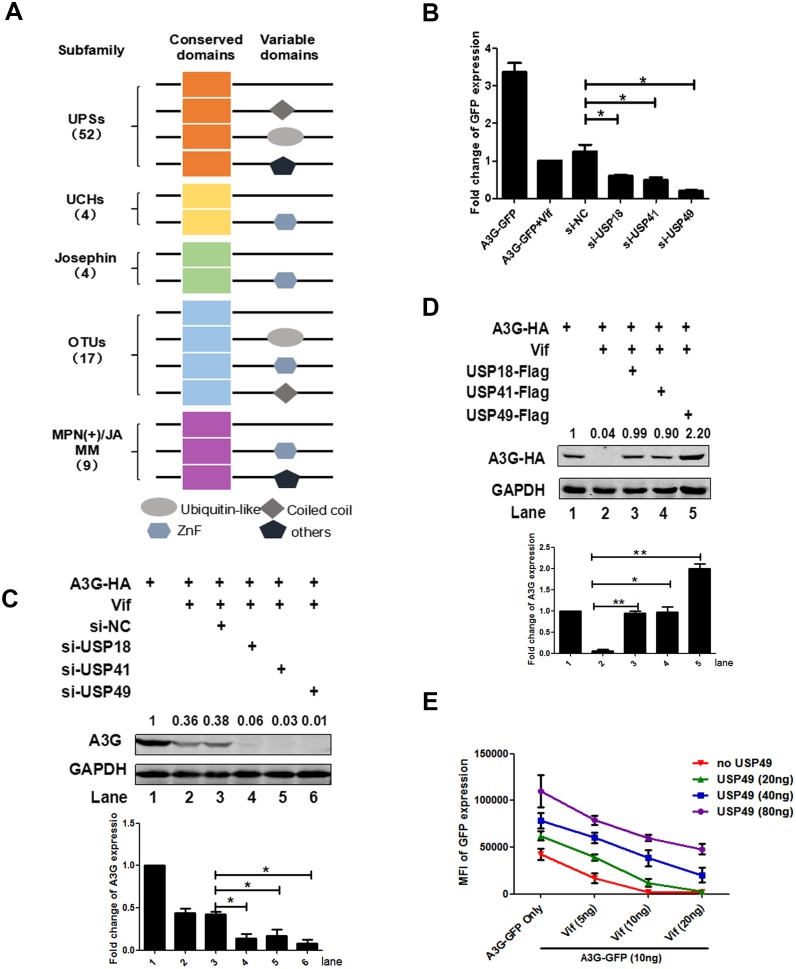
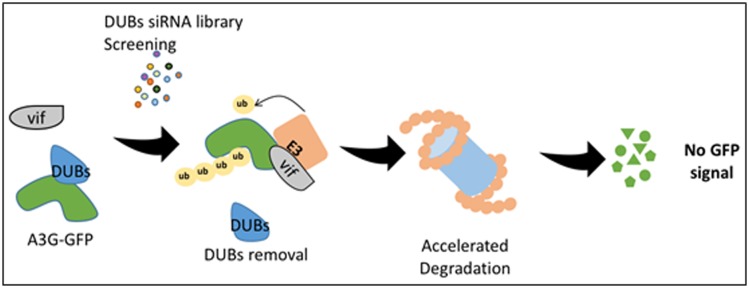
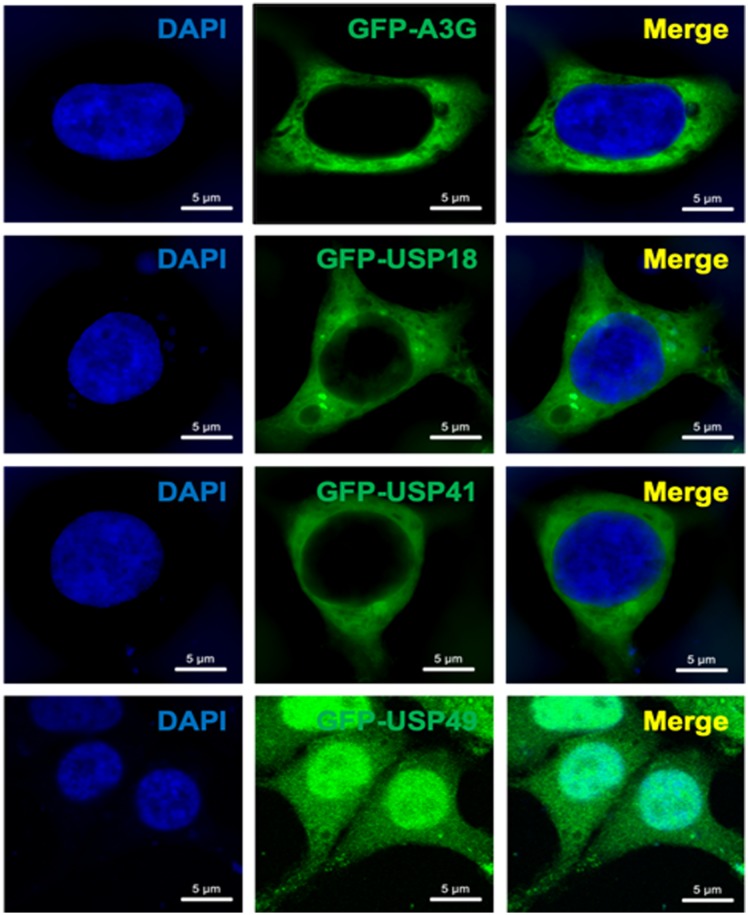
Figure 1. Screening of a DUB-RNAi library identifies that USP 18/41/49 regulates the expression of A3G.
(A) Domain organization of five DUB subfamilies. (B) HEK293T cells were seeded into a 96-well plate with 20,000 cells/well, and then transfected with plasmids expressing A3G-GFP and Vif-HA, as well as siRNAs specific for USP18, USP41, or USP49 respectively. The GFP expression was detected with a PE Envision at 48 hr post-transfection. Error bars represent the SEM of three independent experiments.* p<0.05. (C) HEK293T cells were transfected with plasmids expressing A3G-HA and Vif-HA, as well as siRNAs specific for USP18, USP41, or USP49 respectively. After 48 hr, cells were lysed and Western blot was performed with the indicated antibodies. Representative data were shown and plotted with at least three independent experiments.* p<0.05. (D) HEK293T cells were transfected with plasmids expressing A3G-HA, Vif-HA, and one of plasmid expressing USP18-Flag, USP41-Flag, USP49-Flag. After 48 hr, cells were lysed and Western blot was performed with the indicated antibodies. Representative data were shown and plotted with at least three independent experiments.* p<0.05, **p<0.01. (E) HEK293T cells were transfected with indicated amounts of plasmids expressing A3G-GFP, Vif-HA, or USP49-Flag. The GFP expression was detected with a PE Envision at 48 hr post-transfection. Error bars represent the SEM of three independent experiments.