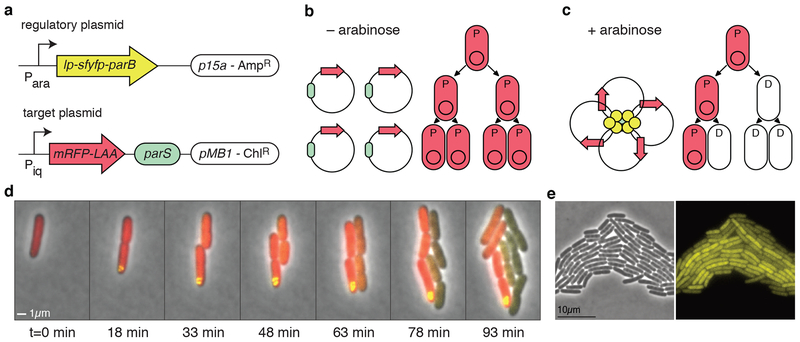
Figure 1.
Asymmetric plasmid partitioning in E. coli. (a) The APP network consists of two plasmids: the regulatory plasmid containing lp-sfyfp-parB (lp: leader peptide) under the control of an arabinose inducible promoter and the target plasmid containing the parS sequence and a red fluorescent protein gene. (b) In the absence of arabinose, there is no expression of lp-sfyfp-parB, so target plasmids are free to diffuse in the cell. This means that they segregate roughly symmetrically in the population in which all cells are progenitor cells (denoted by “P”). (c) When arabinose is present, lp-sfYFP-ParB binds parS on the target plasmid, forming a nucleoprotein complex that gathers all copies of the plasmid together. In this case, cells begin to asymmetrically divide, giving rise to differentiated cells (denoted by “D”). (d) Time-lapse fluorescence microscopy of cells undergoing asymmetric plasmid partitioning. Shown at time t=0 is a single progenitor cell that is in the presence of arabinose. A nucleoprotein complex quickly forms (yellow punctum), and subsequent daughter cells lose the target plasmid. The inherited red fluorescent protein in the daughter cells quickly decays through dilution and proteolysis. (e) Phase contrast (left) and yellow fluorescence microscope images of cells containing only the regulatory plasmid, encoding lp-sfyfp-parB, induced with 0.2% arabinose.