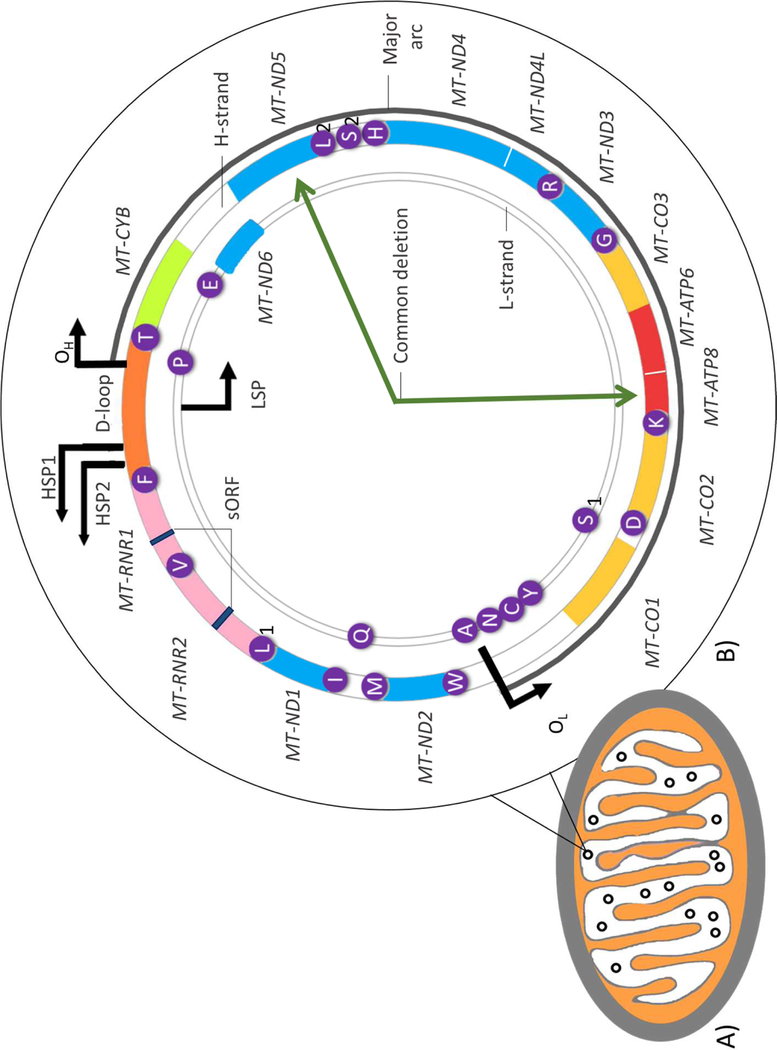
Figure 1. The mitochondrion and structure of human mitochondrial DNA.
A). Basic structure of a mitochondrion containing multiple copies of mitochondrial DNA (mtDNA) represented as black circles. B) Structure of mtDNA: a 16 569 bp circular, double-stranded molecule consisting of a guanine-rich heavy strand (H-strand) and a light strand (L-strand), rich in cytosine. Approximately 93% of the genome is coding, having only one significant non-coding control region which includes the displacement loop (D-loop). This stretch of DNA contains the origin of H-strand replication (OH) the H-strand transcriptional promoters (HSP1 and HSP2) as well as the L-strand promoter (LSP). The DNA 37 primarily contiguous genes, nine on the L-strand and 28 on the H-strand. Twenty-four genes encode mature RNA products: one 12 s rRNA (small ribosomal subunit) and one 16 s rRNA (large ribosomal subunit), and 22 mitochondrial tRNAs (genes highlighted in purple). The remaining 13 mtDNA genes encode polypeptide components of the electron transport chain (ETC) involved in energy production via oxidative phosphorylation (OXPHOS). These include seven subunits (MT-ND1,2,3,4,4L,5,6) of complex I, one subunit (MT-CYB) of complex III, three subunits (MT-CO1,2, and 3) of complex IV, and two subunits (MT-ATP6 and 8) of complex V. mtDNA also encodes two mitochondrial-derived peptides, MOTS-c and Humanin, as short open reading frames (sORF) in the MT-RNR1 and MT-RNR2 genes respectively. Deletions in mtDNA frequently occur in the major arc (indicated by the dark grey line) between the heavy and light strand origins of replication (OH and OL respectively). Among these deletions is the ‘common deletion’: a 4 977-base-pair deletion located between the MT-ATP8 and MT-ND5 genes as indicated by the dark green arrows.