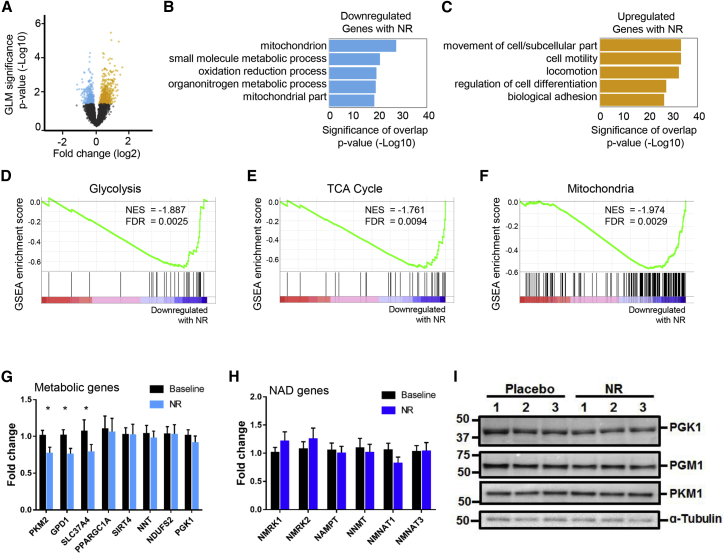
Figure 2.
NR Supplementation Induces a Transcriptional Signature in Human Skeletal Muscle
(A) Differential gene expression analysis on baseline and NR-treated muscle samples (n = 12 at each phase). Volcano plot of differential gene expression between baseline and NR treated human muscle samples. Fold change (Log2, x axis) of gene expression is plotted against p value for differential gene expression (–Log10, y axis). Colored dots represent Ensembl genes that are either upregulated (in orange) or downregulated (in blue) upon NR supplementation at a p value < 0.05.
(B and C) Gene Ontology analysis of significantly dysregulated genes upon NR supplementation for (B) downregulated genes and (C) upregulated genes. Gene Ontology analysis was performed using GSEA. Bars represent the p value (–Log10) of overlap from hypergeometric distribution.
(D) Gene set enrichment analysis (GSEA) suggests that genes belonging to the gene set “glycolysis” are downregulated upon NR supplementation. The normalized enrichment score (NES) and nominal p value are presented on the top-left corner of the graph.
(E) As in (D), but for genes involved in the TCA cycle.
(F) As in (D), but for genes involved in the gene set “mitochondria.”
(G) A qPCR analysis of a select panel of downregulated genes identified through differential gene expression analysis. GAPDH was used as housekeeping gene. Error bars represent SEM (n = 12).
(H) As in (G), but for NAD+ pathway-related genes.
(I) Quantification of phosphoglycerate kinase 1 (PGK1), phosphoglucomutase 1 (PGM1), and pyruvate kinase M1 (PKM1) proteins using immunoblotting assay. Tubulin was used as a loading control.
Data are obtained from 12 participants at each phase and wherever relevant are presented as mean ± SEM. Significance was set at p < 0.05. The absence of significance symbols indicates a lack of statistical significance.