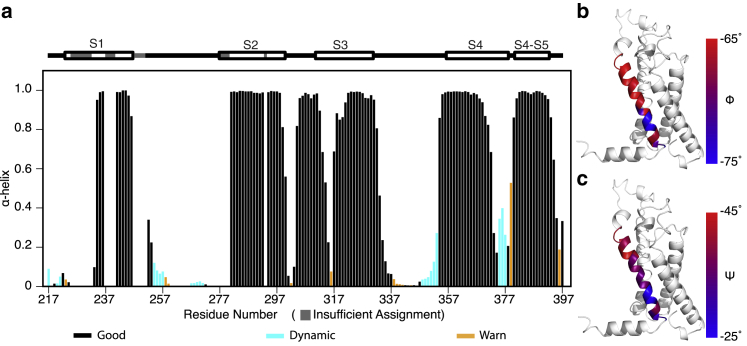
Figure 2.
Secondary structure and backbone dihedral angles of the isolated Shaker-VSD. TALOS+ analysis of the backbone NMR chemical shifts (HN, N, Co, Cα, and Cβ) can reliably predict the backbone dihedral angles and secondary structure elements in proteins. The α-helical propensity of the isolated Shaker-VSD under the conditions used for NMR studies is shown in (a), with numbering according to the full-length Shaker sequence. The expected secondary structure is shown above the data with lines and helices designating loops and rectangles, respectively, and regions with insufficient chemical shift assignments for TALOS+ analysis are drawn in gray. For the predicted α-helical propensity, bars are colored according to the quality of prediction indicated by TALOS+, with black indicating good prediction, cyan indicating dynamic conformations, and orange indicating ambiguous prediction. The data correspond quite well with the expected helical segments. The backbone dihedral angles, φ (b) and ψ (c), of the S4 transmembrane helix are plotted on the homology model of isolated Shaker-VSD with a blue to red color scale to illustrate that the reduced α-helical propensity in the C-terminal half of S4 is due to a transition to 310-helix. Backbone dihedral angles characteristic of α-helix are colored red, and torsion angles characteristic of 310-helix are colored blue in both (b) and (c).