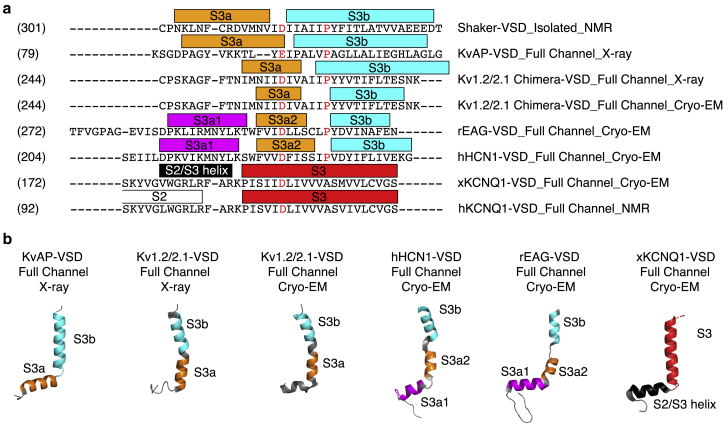
Figure 5.
Diversified secondary structure configuration of S2-S3 linker and S3 helix in different Kv-VSDs. (a) Sequence alignment of the S2-S3 linker and S3 helix region of different Kv-VSDs is shown. Colored rectangles above the sequences indicate the segments corresponding to S3a1 (magenta), S3a2 (orange), S3b (cyan), straight S3 helix (blue), S2-S3 helix (gray), or S2 helix (white). Backbone φ and ψ angles extracted from the Protein Data Bank structures were extracted and analyzed (Ramachandran plots in Fig. S10) to define the helical regions: isolated Shaker-VSD (our NMR results); KvAP-VSD in full channel, x-ray (1ORQ); Kv1.2/2.1-VSD in full channel, x-ray (2R9R); Kv1.2/2.1-VSD in full channel, cryo-EM (6EBK); human KCNQ1-VSD, NMR; X. laevis KCNQ1-VSD in full channel, cryo-EM (5VMS); EAG-VSD in full channel, cryo-EM (5K7L); and HCN1-VSD in full channel, cryo-EM (5U6O). (b) The defined helical regions are shown in the same color scheme plotted on the segment of the structure corresponding to the sequence alignment in (a) for each of the different Kv-VSDs, highlighting the structural diversity in this region. Comparison of the variable S3 helix (also Figs. S5 and S11) suggests that the length and position of helices within this region in Shaker-VSD might be more similar to KvAP.