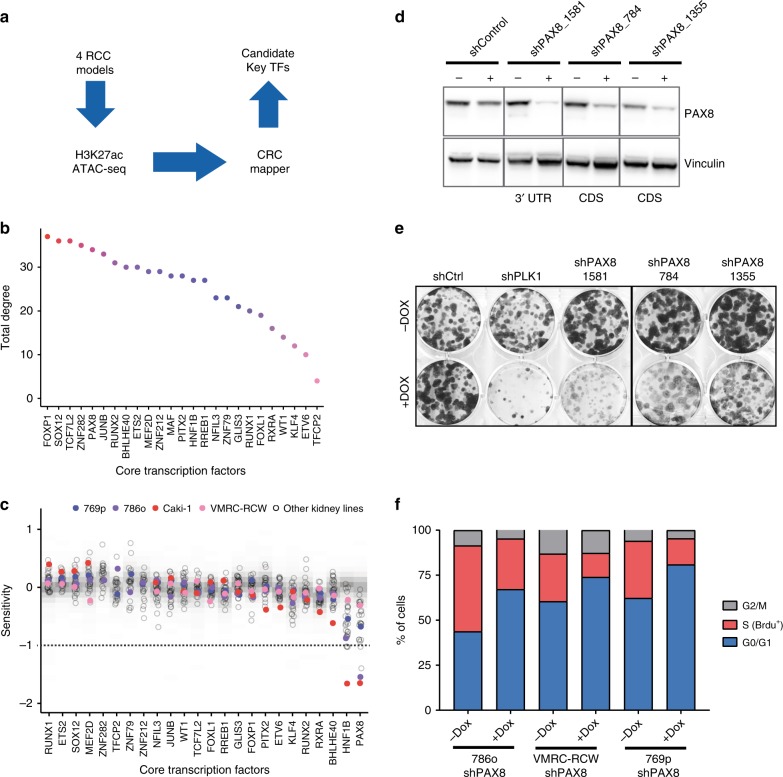
Fig. 1.
Core regulatory circuitry analysis identified PAX8 as a candidate oncogene in RCC. a Diagram of the experimental/computational pipeline to identify critical TFs in RCC. b CRC mapper output as TFs (x-axis) ranked by total degree (y-axis) in RCC. Total degree is a measure of a TFs’ contribution based on how often a given TF can participate in a regulatory interaction with the other TFs. c Sensitivity profile for CRC TFs (x-axis) in combined shRNA datasets (DRIVE, Achilles, Marcotte), where smaller values indicate that the cell line is more dependent on a given gene. Empty dots represent the sensitivity score for each TF in a RCC cell line. Colored dots represent the cell lines originally used in the CRC mapping. Shaded gray represents the distribution of sensitivity values for each TF in cell lines from other lineages. Horizontal dashed line at −1 represents the threshold for significant dependency. d Western blot validation of PAX8 knockdown efficacy in 786o cells infected with dox-inducible shRNAs. e Colony formation assay of 786o cells bearing various doxycycline-inducible shRNAs performed during 10 days. PLK1 is an essential gene in 786o cells and it is a positive control in this assay. f Cell cycle analysis by flow cytometry using PI-BrdU labeling in RCC cell lines bearing shPAX8_1581 upon 4 days of treatment with doxycycline