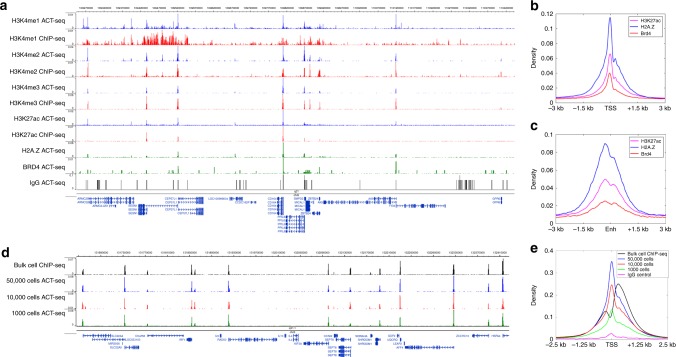
Fig. 1.
ACT-seq robustly maps epigenetic marks in bulk-cell samples. a Genome browser image depicting enrichment of the indicated epigenetic factors in HEK293T cells at a representative genomic region. Data were obtained using ACT-seq (blue, green) or ChIP-seq (red). The ChIP-seq samples were obtained from published ENCODE data sets. A mock IgG sample (aggregated from all ACT-seq IgG replicates) is included as a comparative control for enrichment. b Metagene profile of average H3K27ac, H2A.Z, and Brd4 enrichment at the transcription start site (TSS) region of annotated genes from the hg19 genome. c Metagene profile of average H3K27ac, H2A.Z, and Brd4 enrichment at enhancer (Enh) regions. Enhancers were identified as regions enriched for H3K27ac that did not overlap with an annotated TSS. d Genome browser image depicting enrichment of H3K4me3 in HEK293T samples of the indicated cell number at a representative genomic region. A published ChIP-seq sample from ENCODE is provided for comparison. e Metagene profile of average H3K4me3 enrichment at the TSS region of annotated genes from the hg19 genome. Samples were obtained using the indicated number of cells. A published ChIP-seq sample from ENCODE is provided for comparison