Abstract
The dengue virus (DENV) is a vital global public health issue. The 2014 dengue
epidemic in Guangzhou, China, caused approximately 40,000 cases of infection and
five deaths. We carried out a comprehensive investigation aimed at identifying the
transmission sources in this dengue epidemic. To analyze the phylogenetics of the
2014 dengue strains, the envelope (E) gene
sequences from 17 viral strains isolated from 168 dengue patient serum samples were
sequenced and a phylogenetic tree was reconstructed. All 17 strains were serotype I
strains, including 8 genotype I and 9 genotype V strains. Additionally, 6 genotype I
strains that were probably introduced to China from Thailand before 2009 were widely
transmitted in the 2013 and 2014 epidemics, and they continued to circulate until
2015, with one affinis strain being found in Singapore. The other 2 genotype I
strains were introduced from the Malaya Peninsula in 2014. The transmission source
of the 9 genotype V strains was from Malaysia in 2014. DENVs of different serotypes
and genotypes co-circulated in the 2014 dengue outbreak in Guangzhou. Moreover, not
only had DENV been imported to Guangzhou, but it had also been gradually exported,
as the viruses exhibited an enzootic transmission cycle in Guangzhou. 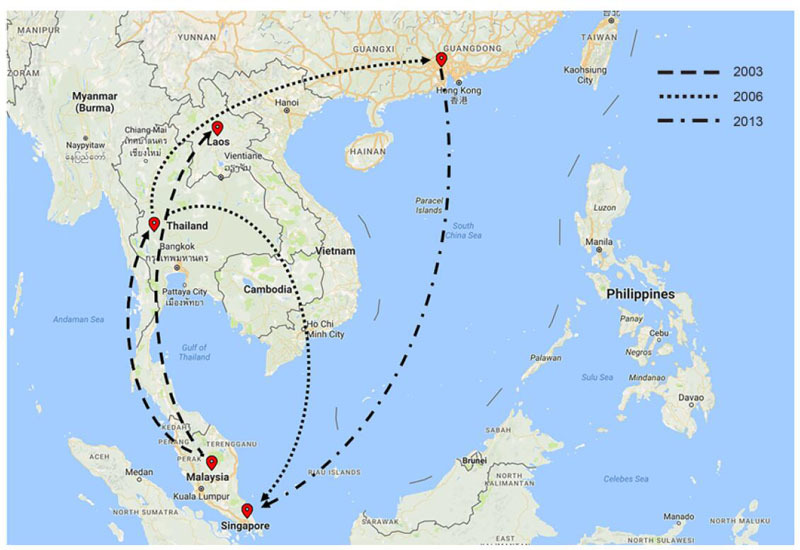
Keywords: dengue virus (DENV), phylogenetic analysis, envelope (E) gene, enzootic transmission cycle
Acknowledgments
This work was supported by the Guangdong Natural Science Foundation (No. S2012030006598), the Science & Technology Planning Project of Guangdong Province of China (No. 2013A020229007), and the Innovative Program of the State Key Laboratory of Virology (No. 2016KF001). We would like to thank the China Center for Type Culture Collection for donating the C6/36 cells, and Jing Peng and Weiyong Liu from the Department of Clinical Laboratory, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, for their guidance on the phylogenetic analyses.
Footnotes
These authors contributed equally to this work.
ORCID: 0000-0002-8326-2895
This article is published with open access at Springerlink.com
Author Contributions
JGW designed and supervised the experiments. GL and PP carried out the experiments. QYH, XJK, KLW and WZ analyzed the data. GL and PP wrote the paper. YTL, HTH, JBL and ZDZ performed sample collections. DW, XHL and XPL provided helpful suggestions about the study. All authors read and approved the final manuscript.
Compliance with Ethics Guidelines
The authors declare that they have no conflict of interest. This study was approved by the Guangdong Province Traditional Chinese Medical Hospital Ethical Review Committee (no. B2015-011-01). Informed oral consent was obtained from all of the patients recruited for the study.
References
- Andrade EH, Figueiredo LB, Vilela AP, Rosa JC, Oliveira JG, Zibaoui HM, Araujo VE, Miranda DP, Ferreira PC, Abrahao JS, Kroon EG. Spatial-temporal co-circulation of dengue virus 1, 2, 3, and 4 associated with coinfection cases in a hyperendemic area of Brazil: a 4-Week Survey. Am J Trop Med Hyg. 2016;94:1080–1084. doi: 10.4269/ajtmh.15-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro P. Evolving RNA virus pandemics: HIV, HCV, Ebola, Dengue, Chikunguya, and now Zika! AIDS Rev. 2016;18:54–55. [PubMed] [Google Scholar]
- Chen R, Vasilakis N. Dengue—quo tu et quo vadis. Viruses. 2011;3:1562–1608. doi: 10.3390/v3091562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Jing Q, Spear RC, Marshall JM, Yang Z, Gong P. Climate and the timing of imported cases as determinants of the dengue outbreak in Guangzhou, 2014: evidence from a mathematical model. PLoS Negl Trop Dis. 2016;10:e0004417. doi: 10.1371/journal.pntd.0004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RL, Voloch CM, Schrago CG. Comparative evolutionary epidemiology of dengue virus serotypes. Infect Genet Evol. 2012;12:309–314. doi: 10.1016/j.meegid.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Cruz CD, Forshey BM, Juarez DS, Guevara C, Leguia M, Kochel TJ, Halsey ES. Molecular epidemiology of American/Asian genotype DENV-2 in Peru. Infect Genet Evol. 2013;18:220–228. doi: 10.1016/j.meegid.2013.04.029. [DOI] [PubMed] [Google Scholar]
- Ditsuwan T, Liabsuetrakul T, Chongsuvivatwong V, Thammapalo S, McNeil E. Assessing the spreading patterns of dengue infection and chikungunya fever outbreaks in lower southern Thailand using a geographic information system. Ann Epidemiol. 2011;21:253–261. doi: 10.1016/j.annepidem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Drumond BP, Mondini A, Schmidt DJ, Bosch I, Nogueira ML. Population dynamics of DENV-1 genotype V in Brazil is characterized by co-circulation and strain/lineage replacement. Arch Virol. 2012;157:2061–2073. doi: 10.1007/s00705-012-1393-9. [DOI] [PubMed] [Google Scholar]
- Ebi KL, Nealon J. Dengue in a changing climate. Environ Res. 2016;151:115–123. doi: 10.1016/j.envres.2016.07.026. [DOI] [PubMed] [Google Scholar]
- Harenberg A, de Montfort A, Jantet-Blaudez F, Bonaparte M, Boudet F, Saville M, Jackson N, Guy B. Cytokine profile of children hospitalized with virologically-confirmed dengue during two phase III vaccine efficacy trials. PLoS Negl Trop Dis. 2016;10:e0004830. doi: 10.1371/journal.pntd.0004830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Burch SS. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000;8:74–77. doi: 10.1016/S0966-842X(99)01669-8. [DOI] [PubMed] [Google Scholar]
- Luo L, Liang HY, Jing QL, He P, Yuan J, Di B, Bai ZJ, Wang YL, Zheng XL, Yang ZC. Molecular characterization of the envelope gene of dengue virus type 3 newly isolated in Guangzhou, China, during 2009–2010. Int J Infect Dis. 2013;17:e498–e504. doi: 10.1016/j.ijid.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Mir D, Romero H, Fagundes DCL, Bello G. Spatiotemporal dynamics of DENV-2 Asian-American genotype lineages in the Americas. PLoS One. 2014;9:e98519. doi: 10.1371/journal.pone.0098519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quam MB, Sessions O, Kamaraj US, Rocklov J, Wilder-Smith A. Dissecting Japan's Dengue Outbreak in 2014. Am J Trop Med Hyg. 2016;94:409–412. doi: 10.4269/ajtmh.15-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke EG, Gregory CJ, Kintziger KW, Sauber-Schatz EK, Hunsperger EA, Gallagher GR, Barber JM, Biggerstaff BJ, Stanek DR, Tomashek KM, Blackmore CG. Dengue outbreak in Key West, Florida, USA, 2009. Emerg Infect Dis. 2012;18:135–137. doi: 10.3201/eid1801.110130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenhuis-Zybert IA, Wilschut J, Smit JM. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Mol Life Sci. 2010;67:2773–2786. doi: 10.1007/s00018-010-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani A, Suzilah I, Malinda M, Anuar I, Mohd MI, Salmah MM, Topek O, Tanrang Y, Ooi SC, Rozilawati H, Lee HL. Aedes larval population dynamics and risk for dengue epidemics in Malaysia. Trop Biomed. 2011;28:237–248. [PubMed] [Google Scholar]
- Sang S, Chen B, Wu H, Yang Z, Di B, Wang L, Tao X, Liu X, Liu Q. Dengue is still an imported disease in China: a case study in Guangzhou. Infect Genet Evol. 2015;32:178–190. doi: 10.1016/j.meegid.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Sang S, Gu S, Bi P, Yang W, Yang Z, Xu L, Yang J, Liu X, Jiang T, Wu H, Chu C, Liu Q. Predicting unprecedented dengue outbreak using imported cases and climatic factors in Guangzhou, 2014. PLoS Negl Trop Dis. 2015;9:e0003808. doi: 10.1371/journal.pntd.0003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfraz MS, Tripathi NK, Faruque FS, Bajwa UI, Kitamoto A, Souris M. Mapping urban and peri-urban breeding habitats of Aedes mosquitoes using a fuzzy analytical hierarchical process based on climatic and physical parameters. Geospat Health. 2014;8:S685–S697. doi: 10.4081/gh.2014.297. [DOI] [PubMed] [Google Scholar]
- Villabona-Arenas CJ, Zanotto PM. Worldwide spread of dengue virus type 1. PLoS One. 2013;8:e62649. doi: 10.1371/journal.pone.0062649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li Y, Feng Y, Zhou H, Liang Y, Dai J, Qin W, Hu Y, Wang Y, Zhang L, Baloch Z, Yang H, Xia X. Phylogenetic analysis of dengue virus reveals the high relatedness between imported and local strains during the 2013 dengue outbreak in Yunnan, China: a retrospective analysis. BMC Infect Dis. 2015;15:142. doi: 10.1186/s12879-015-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. 2009. [PubMed] [Google Scholar]
- Wu W, Bai Z, Zhou H, Tu Z, Fang M, Tang B, Liu J, Liu L, Liu J, Chen W. Molecular epidemiology of dengue viruses in southern China from 1978 to 2006. Virol J. 2011;8:322. doi: 10.1186/1743-422X-8-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HM, Boldrini JL, Fassoni AC, Freitas LF, Gomez MC, de Lima KK, Andrade VR, Freitas AR. Fitting the incidence data from the city of Campinas, Brazil, based on dengue transmission modellings considering time-dependent entomological parameters. PLoS One. 2016;11:e0152186. doi: 10.1371/journal.pone.0152186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Lu L, Fu G, Zhong S, Ding G, Xu R, Zhu G, Shi N, Fan F, Liu Q. Epidemiology and vector efficiency during a dengue fever outbreak in Cixi, Zhejiang Province, China. J Vector Ecol. 2009;34:148–154. doi: 10.1111/j.1948-7134.2009.00018.x. [DOI] [PubMed] [Google Scholar]
- Yong YK, Thayan R, Chong HT, Tan CT, Sekaran SD. Rapid detection and serotyping of dengue virus by multiplex RT-PCR and real-time SYBR green RT-PCR. Singapore Med J. 2007;48:662–668. [PubMed] [Google Scholar]
- Zellweger RM, Prestwood TR, Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe. 2010;7:128–139. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhang FC, Zhu Q, Wang J, Hong WX, Zhao LZ, Deng YQ, Qiu S, Zhang Y, Cai WP, Cao WC, Qin CF. Epidemiological and virological characterizations of the 2014 dengue outbreak in Guangzhou, China. PLoS One. 2016;11:e0156548. doi: 10.1371/journal.pone.0156548. [DOI] [PMC free article] [PubMed] [Google Scholar]


