Abstract
Machine learning (ML) algorithms have found increasing utility in the medical imaging field and numerous applications in the analysis of digital biomarkers within positron emission tomography (PET) imaging have emerged. Interest in the use of artificial intelligence in PET imaging for the study of neurodegenerative diseases and oncology stems from the potential for such techniques to streamline decision support for physicians providing early and accurate diagnosis and allowing personalized treatment regimens. In this review, the use of ML to improve PET image acquisition and reconstruction is presented, along with an overview of its applications in the analysis of PET images for the study of Alzheimer's disease and oncology.
Keywords: molecular imaging of neurodegenerative diseases, cancer detection imaging, PET, Alzheimer's, cancer imaging
Introduction
Machine learning (ML) algorithms have emerged as critical infrastructure in the analysis of digital biomarkers within positron emission tomography (PET) imaging. The application of ML to nuclear imaging is multidisciplinary in nature, drawing upon expertise from the computer sciences, statistics, and the physics and chemistry of nuclear imaging, in order to address unmet challenges in the treatment of neurodegenerative diseases and oncology. The number of publications that have discussed the use of ML algorithms in PET has grown exponentially in the past decade (Figure 1). The use of ML is especially prevalent in the field of radiomics, where advanced algorithms analyze medical images and extract subtle features that are diagnostic of disease.1–3 Large datasets of digital medical information are particularly suitable toward these analysis methods, although limited patient datasets have also been applied to ML algorithms, typically in proof-of-principle type studies. The use of artificial intelligence in the study of neurodegenerative diseases and oncology has the potential to significantly reduce the time required for radiologists to interpret PET scans and provide decision support to physicians diagnosing patients and selecting personalized treatment regimens. Despite their well-recognized utility, several challenges of ML algorithms must be considered prior to their successful implementation in PET image analyses.4–7
Figure 1.
Number of publications in PubMed per year (from January 1995 to April 2019) using the keywords “deep learning” or “machine learning” and “PET”.
This review presents an overview of the recent applications of ML in the areas of Alzheimer's disease (AD) and oncology, as well as its utility in improving the acquisition and reconstruction of PET images. Given that several review articles are available regarding the use of ML in the diagnosis and prognosis of AD,8 as well as in oncology,9 this article primarily focuses on surveying selected recent work.10 Electronic search results from the PubMed database used a combination of the search terms “Deep Learning OR Machine Learning OR Artificial Intelligence AND PET”. No start time was used, articles were however selected for review up until which point it superimposed with previous reviews. Search results were extended up until April 1, 2019. Additional relevant articles were selected manually using the cited references from the articles that appeared in the search results. Inclusion criteria included research articles or abstracts that incorporated ML into any aspect of PET imaging acquisition and analysis. Exclusion criteria included articles not in English and those not fully available. A total of 43 articles were included for this review. Furthermore, readers interested in a tutorial on the fundamental principles of ML are directed toward a recent article by Uribe et al.,11 as it is covered only briefly in this review.
Principles of ML
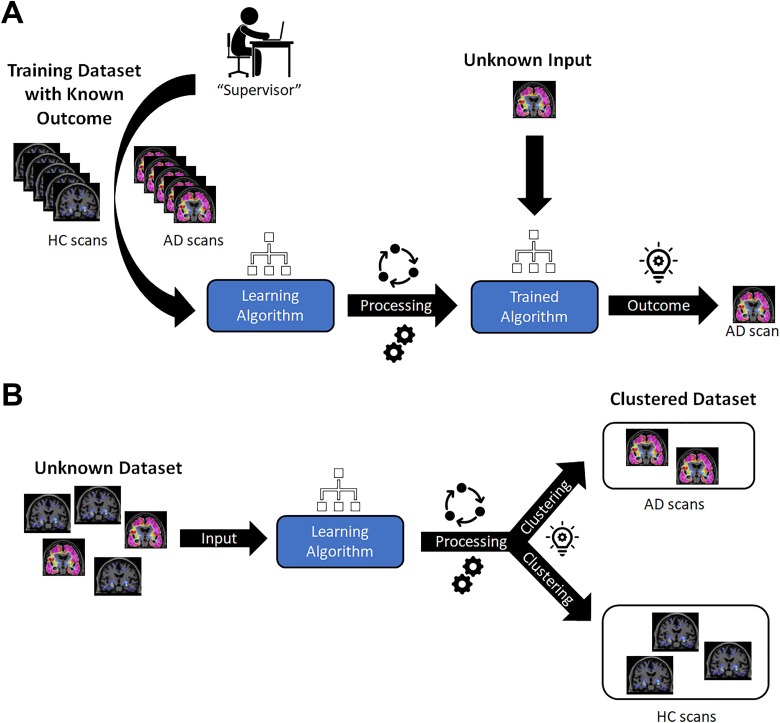
Machine learning algorithms may be categorized as either supervised or unsupervised, both of which have been described in detail elsewhere.9,11 Briefly, supervised learning involves a data set being provided to an algorithm, or neural network, which then aims to find a mathematical function that can map the input data into output labels, ultimately mapping the connection between a given set of input variables and the desired output parameters (Figure 2A). Thus, the algorithm must be provided with an appropriate and representative design space to generate adequate classification and predictive capabilities.12 The ML algorithms employed in PET imaging studies typically perform supervised learning using artificial neural networks (ANNs)—wherein convolutional neural networks (CNNs) have become an increasingly popular subset—as well as random forests.11 In unsupervised learning, an algorithm is applied to a data set for which the output is unknown. A common goal for unsupervised algorithms is to find commonalities within a data set, for instance, differentiating between a disease and non-disease state for which no prior knowledge has been provided (Figure 2B). A common unsupervised algorithm involves data clustering, including K-means clustering and hierarchical clustering, as previously described.11
Figure 2.
Schematic representations of ML algorithms tasked to differentiate individuals diagnosed with AD from HCs. A, An illustration of supervised learning. The ML algorithm would be provided with a cross-sectional data set of PET brain images that contain examples of all subgroups, and be told in advance of the classification, so as to learn the distinguishing features. The appropriately mapped algorithm would ideally be able to provide a classification of AD or HC, provided the image is within the design space in which it learned. B, An illustration of unsupervised learning. The ML algorithm would be provided with a cross-sectional data set of PET brain images, but not be told in advance of the classification. The ML algorithm would then find commonalities so as to cluster the data into homogeneous subgroups. AD indicates Alzheimer's disease; HCs, healthy controls; ML, machine learning; PET, positron emission tomography.
Standardized radiomic datasets are required for the successful implementation of ML algorithms. In recent years, publicly available standardized databases of PET brain scans of patients with AD have been compiled with databases such as the Alzheimer’s Disease Neuroimaging Initiative (ADNI), the Harvard Aging Brain Study (HABS), Australian Imaging, Biomarker & Lifestyle Flagship Study of Aging (AIBL), the Dominantly Inherited Alzheimer's Network (DIAN), Biomarkers for Identifying Neurodegenerative Disorders Early and Reliably (BioFINDER), Development of Screening Guidelines and Criteria for Predementia Alzheimer’s Disease (DESCRIPA), and Imaging Dementia—Evidence for Amyloid Scanning (IDEAS). Many of these databases have been made publicly available from multicenter clinical trials in patients ranging from cognitively normal to AD, using predominantly [18F]fluorodeoxyglucose ([18F]FDG) to image glucose metabolism, as well as amyloid plaque imaging agents, namely [11C]Pittsburgh compound B ([11C]PiB) and the FDA-approved radiotracers Amyvid ([18F]florbetapir or [18F]FBP), Neuraceq ([18F]florbetaben or [18F]FBB), and Vizamyl ([18F]flutemetamol or [18F]FLUT).13 As with the AD datasets, the most common radiotracer in oncology is [18F]FDG; however, there are far fewer large PET scan databases in oncology. Two of note are the Web-Based Imaging Diagnosis by Expert Network (WIDEN) and The Cancer Imaging Archive (TCIA), both of which continue to grow with ongoing contributions which should ultimately lead to an increase in studies involving ML in oncology.14–16
Improving PET Acquisition and Reconstruction With ML
Photon Attenuation Correction
Photon attenuation, defined as the loss of photon flux intensity through a medium, remains a significant challenge when obtaining high-resolution PET images. Attenuation is typically due to unavoidable factors such as interaction of the photon within the body tissue prior to detection. This oftentimes results in the loss of image resolution, formation of artifacts, and distortion of images, and is particularly a problem for larger individuals. Attempts to correct for attenuation are done so on an individualized basis via the generation of attenuation maps, which occur during the image processing stage of the data after it has been collected. Attenuation maps are most accurately generated using scaling from the images of an accompanying computed tomography (CT) scan as this can account for the attenuation caused by bone. This is not possible from magnetic resonance (MR) scans due to the lack of signal contrast between bone and air.17 Accurate attenuation maps are thus challenging to obtain for PET/MR scans or PET imaged alone, as a direct measure of photon attenuation is not recorded. An inaccurate MR-based attenuation correction (MR-AC) may also result in large underestimations of radiotracer quantity in patients with cancer, depending on the proximity of the tumors to bone.18,19 As the use of CT may also not be appropriate for all individuals, for instance, with pediatric patients, recent studies have utilized ML algorithms to address the issue of attenuation correction (AC), with the aim of obtaining accurate attenuation maps from solely PET/MR data or PET alone as the input modalities.20–22
Liu et al. recently developed an ML algorithm (named deepAC) to automate the PET image AC process without input from a separate anatomical image such as CT.21 The model was first trained using [18F]FDG PET/CT datasets. This subsequently allowed for the generation and optimization of continuously valued “pseudo-CT” images solely from the uncorrected [18F]FDG images that were used to generate the attenuation map. The resulting attenuation-corrected [18F]FDG image generated by deepAC was found to be quantitatively accurate with average errors of less than 1%. Similarly, Ladefoged et al.22 implemented an ML method for PET/MR-AC (named DeepUTE) designed for children with suspected brain tumors. Using an AC method derived from CT (CT-AC) serving as reference standard, the authors evaluated the DeepUTE method relative to the results of an MR-derived AC method termed RESOLUTE, which had been previously reported by the same investigators.23 The authors found that while the AC generated from both the RESOLUTE and DeepUTE methods reproduced the CT-AC reference to a clinically acceptable precision, DeepUTE more robustly reproduced the standard on both qualitative inspection and quantitative evaluation.
Obtaining an accurate AC for PET images of other regions of the body (whole-body AC) has limited accuracy and is considered more challenging than those for PET images of the brain. Whereas brain images utilize templates/atlases to provide a generic reference standard with which to compare, such templates are not available nor practical for whole-body PET scans. Torrado-Carvajal et al. proposed a Dixon-VIBE (Volumetric Interpolated Breath-Hold Examination) ML algorithm (named DIVIDE) to help produce pseudo-CT maps of the pelvis based only on standard Dixon-VIBE MR images.24 The ability to map between a CT and MR image slice allows for a more accurate AC to be generated from MR data, thus offering an alternative method to produce an accurate AC for combined PET/MR scanners. Hwang et al. similarly developed a new ML CNN-based approach using whole-body [18F]FDG PET/CT scan data of patients with cancer (n = 100) for training and testing.25 The method utilized the maximum likelihood reconstruction of activity and attenuation algorithm as inputs to produce a CT-derived attenuation map and compared the resulting AC to those obtained using Dixon-MR images, using the CT-AC as reference standard. The method produced less noisy attenuation maps and achieved better bone identification for whole-body PET/MR studies relative to the Dixon-based 4-segment method currently used. Bradshaw et al. also evaluated the feasibility of a 3-D CNN (deepMRAC) to produce MR-AC from combined PET/MR images of the pelvis for patients with cervical cancer.26 DeepMRAC was trained using reference CT image data in order to simulate a pseudo-CT that discretized different tissue zones (fat/water, bone, and air) and served to produce a CT-based AC from PET/MR data. The results showed that the distribution of errors for the standardized uptake value (SUV) of cervical cancer lesions was significantly narrower using the ML-based MR-AC compared to the standard Dixon-based MR-AC method.26
PET Image Reconstruction
Several techniques currently exist to reconstruct PET images from tomographic PET sinogram data. Conventional approaches include analytical filtered back projection and iterative maximum likelihood methods like maximum likelihood expectation maximization (MLEM) and ordered subset expectation maximization (OSEM). Häggström et al. developed a convolutional encoder–decoder network, named DeepPET, which served to produce quantitative PET images using sinogram data as input.27 DeepPET was found to produce higher quality images relative to the filtered back projection and OSEM methods based on such metrics as relative root mean squared error (lower by 53% and 11%, respectively), structural similarity index (higher by 11% and 1%, respectively), and peak signal to noise ratio (higher by 3.8 and 1.1 dB, respectively). DeepPET also produced images 3 and 108 times faster than filtered back projection and OSEM methods, respectively, hence demonstrating a reduction in the computational cost for image reconstruction.
Artificial neural networks have also been used at the PET image reconstruction stage to improve the resolution of the PET images generated from the raw datasets. Conventional reconstruction methods such as the iterative expectation maximization algorithm lead to increased noise. Alternatively, maximum a posteriori (MAP) estimates reduce divergence at higher iterations, but total variation MAP (TV-MAP) image reconstruction may lead to blurring. Yang et al. proposed a patch-based image enhancement scheme, using an ANN model called multilayer perception (MLP) with backpropagation to enhance the MAP reconstructed PET images.28 The MLP model was trained using image patches that were reconstructed using the MAP algorithm. The MLP method produced images with reduced noise compared to MAP reconstruction algorithm, resulting in a reduction in size of the unachievable region. Similarly, Tang et al. employed dictionary learning (DL)-based sparse signal representation for MAP reconstructed PET images.29 The employed DL-MAP algorithm trained on the corresponding PET/MR structural images, and the results were compared to those from conventional MAP, TV-MAP, and patch-based algorithms. The DL-MAP algorithm produced images with improved bias and contrast, with comparable noise to those produced using other MAP algorithms. Liu et al. recently developed an image reconstruction algorithm using ML neural networks, consisting of 3 modified U-Nets (3U-Net), to improve the signal to noise ratio of PET images obtained from multimodal PET/MR image data, without the need for a high-dose PET image. The 3U-Net model using PET/MR data as input produced a reconstructed PET image with improved signal to noise ratio relative to those using PET input data alone or PET/MR in a 1U-Net model.30
Spatial Normalization
Spatial normalization (SN) of brain images, namely, the formatting of brain images (initially in pre-normalized or native space) into a standard anatomical space such as Talairach or MNI coordinates, aids greatly in the standardization of PET brain images. The activity within specific pre-defined regions of a PET image upon conversion to a standard anatomical space may be quantified more reliably relative to an image analyzed in native space. Accurate SN of brain PET images is most reliably carried out using the data from accompanying MR scans; hence, obtaining an accurate SN of amyloid PET images without MR remains a challenge. Kang et al. trained 2 CNNs: convolutional autoencoder and generative adversarial network, using a patient dataset (n = 681) of simultaneously acquired [11C]PiB PET/MR scans of individuals with AD, mild cognitive impairment (MCI), and healthy controls (HCs).31 The training regimen produced a spatially normalized PET image, given the transformation parameters obtained by the SN generated from the accompanying MR data. As a result, given an inputted PET image in native space, the neural network was able to generate an individually adaptive amyloid PET template to achieve accurate SN without the use of MR data. The ML method was found to reduce the SN error relative to when an average amyloid PET template is used.
Ultralow Dosage for PET Scans
Although radiotracer dosage requirements during a standard PET or PET/CT image acquisition would not be expected to impart a pharmacological effect on the patient, there has been growing concern regarding the accompanying radioactivity exposure.32 As such, it may at times be desirable to carry out PET imaging using a reduced dosage of radiotracer, particularly in instances where a full dose may not be appropriate, such as with younger populations. The ability to carry out a low-dosage PET scan would also greatly expand the scope of synthetic strategies currently used for radiotracer development. This is particularly true for 11C-labeled tracers, where the relatively short half-life (t 1/2 = 20.3 minutes) prohibits the use of many conventional organic synthesis techniques that could otherwise be utilized for radiotracer preparation. Low-dose PET images are inherently noisy, however, which makes it difficult to draw qualitative and/or quantitative conclusions from the data. To this end, efforts have been made to develop ML methods that would allow reduced radiotracer quantities for PET/MR imaging without sacrificing the diagnostic quality of the image. The application of ML algorithms to address this challenge simulates a low dose by a fraction (ca. 1 - 25%) of acquired PET data from a full-dose image. The ML method then predicts the images of a full-dose using only the low-dosage data as the input.
Chen et al. analyzed one one-hundredth of acquired PET data scans—roughly the radiation exposure equivalent of a transcontinental flight—using the amyloid tracer Neuraceq from simultaneously acquired PET/MR imaging modalities, hence simulating a low-dosage acquisition while still using the full MR data set.33 ML methods were then utilized to predict the full-dose PET image, using the experimental PET/MR as reference standard. The simulated full-dose PET image was found to satisfactorily reproduce the experimentally acquired full-dose image. Similarly, Yang et al. developed a shallow ANN, which served as a learning-based denoising scheme, using image patches from PET scans from 5 individuals as the sample data, and the synaptic vesicle glycoprotein 2A radiotracer, [11C]UCB-J.34 The authors used an MAP reconstruction algorithm as a learning-based denoising scheme to process 3-D image patches from reduced-count PET images, thereby producing simulated full-count reconstruction image patches from the reduced-count data.
Xiang et al. used a CNN model that was adapted to use both low-dose PET (ca. 25% of a full dose) and MR image data as input modalities.35 In order to simulate a full-dose PET image, the algorithm was developed to map the connection between the inputs and the full-dose PET data. The simulated image was further refined by integrating multiple CNN modules following an auto-context strategy, thereby generating a full-dose PET image that was of competitive quality with previously reported simulation algorithms but requiring only ca. 2 seconds of processing time, as opposed to other simulation schemes that required 16 minutes. Similarly, Kaplan et al. created a residual CNN model to estimate a full-dose equivalent PET body image using one-tenth of the input data.36 Particular care was taken in the ML model to preserve edge and structural features in the image by accounting for them during training. The body image was divided into different regions (for instance, brain, heart, liver, and pelvis) and training the model for each region was done so separately, while a low-pass filter was used to denoise the low-dose PET image scans while preserving important structural details. The algorithm produced significantly improved image qualities relative to the input low-dose PET image slice data, generating images that were comparable in quality to the full-dose image slices.
The potential use of ultralow-dose [18F]FDG in the automated detection of lung cancer with an ML algorithm has also been studied.37 An ANN, trained to discriminate patients with lung cancer from HC, was first assessed data from 3936 PET slices exported from images with and without lung tumors. Three separate reconstructions of the data were performed using the standard clinical effective dose (3.74 mSv) relative to simulated low dosages: a 10-fold (0.37 mSv) and 30-fold (0.11 mSv) reduction of the standard effective dose. The investigators report high sensitivity, 91.5%, and high specificity, 94.2%, with the ultralow dose (0.11 mSv), suggesting that applying ML to PET data enables the use of ultralow doses.37
Machine Learning PET Analysis: Applications in AD
Early Diagnosis of AD
PET has become an essential diagnostic tool in AD to improve diagnosis, monitor disease progression, and select patients for clinical trials.38,39 Big data and artificial intelligence methodology has been applied to analyze PET images to help differentiate and predict individuals with MCI who will eventually progress to AD from those who will not.8 [18F]FDG has been shown to illustrate the decline in regional cerebral glucose metabolism related to AD prior to the onset of structural changes, allowing for an earlier diagnosis relative to clinical evaluation.40 Ding et al. developed an ML model to predict the future clinical diagnosis of AD, MCI, or neither, from [18F]FDG imaging data made available via the ADNI database (n = 1002).41 The authors created a CNN of Inception V3 architecture, which was trained on 90% of the data set, while the remaining data set was tested to evaluate the predictive power of the model. The authors were able to demonstrate that the ML algorithm was able to achieve 100% sensitivity at predicting the final clinical diagnosis in the independent test set, an average of 75.8 months (6.3 years) prior to final diagnosis. Lu et al. developed a single-modality ML algorithm using [18F]FDG images of patients (n = 1051; data obtained from the ADNI database) with AD, stable MCI (sMCI), progressive MCI (pMCI) or HC, for training and evaluation.42 The method was able to identify patients with MCI who would later develop AD to an accuracy of 82.51%, thus differentiating between those who exhibited nonprogressive sMCI (median follow-up time of 3 years).
Similarly, Lu et al. developed an ML algorithm for the early diagnosis of AD using multimodal [18F]FDG PET/MR and multiscale deep neural networks.43 The new method first segments the brain image into cortical and subcortical gray matter compartments prior to subdividing each into image patches of hierarchical size, hence extracting features at coarse-to-fine structural scales and preserving structural and metabolism information. The method obtains 82.4% accuracy in identifying individuals with MCI who will convert to AD 3 years prior to conversion, in addition to 94.23% sensitivity in classifying individuals with a clinical diagnosis of probable AD. Choi and Jin developed a CNN that, as opposed to feature-based quantification approaches, did not require a preprocessing of the images such as SN or manually defined feature extraction prior to analysis.44 Data obtained from the ADNI database were used to train and test the CNN model and were comprised of [18F]FDG and Amyvid scans from 492 patients. A prediction accuracy of 84.2% for the conversion of MCI to AD was obtained, outperforming conventional feature-based quantification approaches.
In addition to the use of supervised ML algorithms in longitudinal studies, recent examples have emerged of unsupervised algorithms aimed to predict disease trajectory in patients clinically diagnosed with MCI. Gamberger et al. employed a multilayer clustering algorithm that identified 2 homogeneous subgroups of patients as rapid decliners (n = 240) and slow decliners (n = 184), from a well-characterized data set ([18F]FDG PET/MR and Amyvid PET/MR scans, including follow-up scans after 5 years) of 562 patients with late MCI obtained from the ADNI database.45 A combination of the Alzheimer’s Disease Assessment Scale–Cognitive subscales 11 and 13 (ADAS-cog-11 and ADAS-cog-13) was then identified as classifiers that best correlated with and predicted the rapid decline in the patients with MCI.
Classification of AD
Automated classification of AD has shown promise with ML algorithms that have been trained to use the data from PET brain images to accurately differentiate between individuals who have been clinically diagnosed with AD and MCI from those of HCs.8 Such models typically identify the patterns and extract the discriminate features related to AD and MCI that distinguish it from other groups. Data from PET images have also been combined with those of accompanying voxel-wise gray matter density maps obtained from MR, which is also informative of the onset of AD.43 The use of these models may prove useful for untrained individuals in aiding with diagnosis, potentially reducing the cost and time lines for clinical trials of novel treatments.
Conventional classification algorithms first segment the PET brain image into various regions of interest (ROIs), which may ignore some minor abnormal changes in the brain relevant to AD diagnosis. Liu et al. proposed a 3-D PET image classification method based on a combination of 2-D CNNs and recurrent neural networks (RNNs), which learns the structural features used for classification of AD diagnosis upon partitioning the 3-D image into a set of 2-D image slices.46 The 2-D CNN model was trained on the features within each 2-D image slice, while the RNN was trained to pull out the features between slices. When evaluated on a set of [18F]FDG images (n = 339), the model exhibited a classification performance of 95.3% for AD versus HCs, while 83.9% performance was obtained when differentiating between individuals exhibiting MCI versus HC. Alternatively, Liu et al. also proposed a CNN classification algorithm that is a construct of multiple 3-D-CNNs used on different local 3-D image patches from multiple modalities such as PET and MR.47 The CNN translates each image patch into discriminative features, after which a high-level 2-D CNN collects the high-level features that were learned. The model can automatically learn discriminatory multilevel and multimodal features, is somewhat robust to scale and rotation variations, and does not require segmentation of the image in preprocessing the brain images. Evaluation of the method finds that it achieves a 93.26% accuracy rate for the classification of AD versus HC and 82.95% for differentiating between HC and pMCI diagnosed patients. Furthermore, Zhou et al. developed a novel deep neural network to identify AD that incorporated a 3-stage effective feature learning and fusion framework while using both genetic input data such as single nucleotide polymorphism, a predictor of AD risk, in addition to imaging modalities like PET and MR.48 The ML method incorporates a flexible architecture to be able to learn from heterogeneous datasets with differences in sample size and distributions. The method, when evaluated for AD diagnosis using PET/MR images from the ADNI database (n = 805), exhibited better classification performance relative to the baseline method as well as other previously reported methods.
Conventional methods for the automated quantification of β-amyloid in PET brain images have used composite standardized uptake value ratios (SUVr), the data of which are more variable in longitudinal studies. Gunn and coworkers recently concluded that β-amyloid accumulation in AD progression is the result of heterogeneous regional carrying capacities and can be mathematically modeled using a logistic growth equation.49 From the logistic growth equation, a new biomarker can be derived, called amyloid load (AβL) to quantify β-amyloid levels. Algorithms to determine AβL from β-amyloid scans and assist in the classification of AD from PET images of the brain have been branded as software called AmyloidIQ. From a test sample of 769 Amyvid scans obtained from the ADNI database, AmyloidIQ demonstrated a 46% increase in mean difference in effect sizes for AβL relative to composite SUVr quantification. An equivalent increase in effect size was found for longitudinal data, assessing patients who had a follow-up PET scan after 2 years.50 The authors envision that providing expert knowledge as commercially available software would allow clinical trials to run more efficiently by decreasing trial size, timelines, and cost.
Subsequently, the same authors developed an algorithm, named TauIQ, that was able to partition a Tau PET image into 3 regions: background nonspecific, global (TauL), and local Tau accumulation, thereby providing quantification data on both global and local tau deposition.51 Images from 234 patients obtained from the ADNI database were analyzed, each with [18F]flortaucipir (Tau tracer), Amyvid (β-amyloid tracer), and MR image data. The Amyvid scans were first analyzed using AmyloidIQ to derive the patient-specific point of disease progression prior to use of the TauIQ algorithm.
Machine Learning PET Analysis: Applications in Oncology
In oncology, PET imaging datasets that are employed in ML are typically performed at a single facility, so investigators may not follow standardized guidelines for comparison to datasets from other PET imaging facilities. Multicentered clinical trials require standardized acquisition and reporting of scans, thereby creating an opportunity to apply ML to the large datasets being generated. A tool for image exchange and review was developed called WIDEN for the purpose of performing a multicenter clinical trial for monitoring and tailoring treatment of Hodgkin's lymphoma with real-time review of images of PET/CT scans.52 Interestingly, despite being based on scans collected to study patients with lymphoma, the scans can be used in ML associated with other clinical situations. For example, an algorithm was developed and applied to 650 PET scans that were available through WIDEN for the purpose of automatically identifying and extracting the average SUV of the liver to allow for automatic assessment of image quality or quickly identifying acquisition problems. The proposed model, LIDEA, was able to identify the liver and quantify the SUV with 97.3% sensitivity, with a 98.9% correct detection rate when co-registered with CT scans.14 Oncologic PET scans have also been made available through TCIA, which has accumulated datasets from various imaging modalities.15 PET tomography imaging datasets are being contributed to TCIA, which should ultimately lead to an increase in studies involving ML in oncology.16
Accurate tumor delineation and segmentation is critical in diagnosis and staging of cancer. ML algorithms for automatic delineation and segmentation of tumor volumes could remove operator variability and provide fast decision support for physicians in radiotherapy planning for patients with cancer and can be used for patient stratification and predicting patient outcomes. Tumor delineation defines the gross tumor volume (GTV) and metabolic tumor volume (MTV), which are considered global measurements and do not convey spatial information of the ROI; these are known as first-order radiomic features. Tumor segmentation defines different regions within the tumor microenvironment that arise from differences in characteristics such as vascularization, cell proliferation, and necrosis.53 This involves second-order radiomic features, or textural features, which provide information on the spatial relationships between intensities ≥2 voxels caused by heterogeneous radiotracer distribution.54 Heterogeneity of the tumor microenvironment can be captured by textural features from various imaging modalities to illustrate tumor characteristics that may contribute to poor prognosis or treatment response.55 The utility of radiomic feature analysis from [18F]FDG imaging in providing physicians with decision support in planning treatment regimens for patients with cancer has been shown in a number of studies.56–60 Manual tumor delineation and segmentation are performed by highly trained experts, yet are time-consuming and open to subjectivity. A point of concern arises in that there is great variability in the analysis of textural features that arises from different acquisition modes and reconstruction parameters.61 Indeed, one study showed that variation in SUV determination methodology significantly altered the results of textural analysis in [18F]FDG imaging; this stresses the importance of standard protocols for the acquisition and reconstruction of PET images.62
Berthon et al. used an automatic decision tree-based learning algorithm for advanced image segmentation (ATLAAS) to develop a predictive segmentation model that includes 9 different auto-segmentation methods and automatically selects and applies the most accurate of these methods based on the tumor traits extracted from the PET scans.63 An ATLAAS has also been successfully used for tumor delineation by automatically determining GTV in patients with head and neck cancer.64 Another study demonstrated automatic tumor delineation with an ML method applied to a deep CNN could automatically define GTV in patients with head and neck cancer.65 Blanc-Durand et al. developed an automatic detection and segmentation method using PET scans from patients with glioma with a full 3-D automated approach using a U-Net CNN architecture.66 The radiotracer used in this study was [18F]fluoroethyltyrosine ([18F]FET), an amino acid radiotracer that may be appropriate in the diagnosis of brain tumors since it exhibits lower normal brain uptake, which leads to lower background.67 The investigators reported 100% sensitivity and specificity for tumor detection, having no false positives, while the segmentation resulted in relatively high performance which may be improved with a larger data set applied to algorithm training. The authors further studied [18F]FET in patients with glioma applying unsupervised learning by K-means clustering for the automatic clustering of tumor voxels to examine radiomic features associated with disease progression and patient survival.68
Studies have shown the potential for the application of ML to study textural features, using CNNs and 3-D CNNs to classify nodal metastases in various cancers.69–72 The ability for ML to predict patient outcomes has been demonstrated with Hodgkin's lymphoma, where radiomic features extracted from the mediastinal region were highly predictive of refractory disease, while features extracted from other regions of the body were not.73 Two different studies using ML with a CNN applied to textural features of [18F]FDG scans were able to predict the response of patients with esophageal cancer to chemotherapy with high sensitivity and specificity.74,75 Kirienko et al. developed an algorithm composed of 2 networks, one as the feature “extractor” and another as a “classifier,” and provided evidence that their method could correctly classify patients with lung cancer based on TNM classification (tumor extent, lymph node involvement, and presence of metastases).72,76 In addition to supervised ML algorithms, unsupervised learning has been applied to textural features. One study examined [18F]FDG textural features that would reflect different histological architectures in patients with different cervical cancer subtypes.77 The authors applied a hierarchical clustering algorithm to their data set examining features including first-order SUV and MTV, as well as second-order radiomic features, and concluded that their approach has implications for personalized medicine and prognostic models.77
Summary and Outlook
The applications of ML algorithms in the medical field have developed substantially in recent years, coinciding with the increase in the computing power of modern-day processors. This narrative review provided an overview of the potential applications of ML algorithms in the various aspects of PET imaging, highlighting several areas that have seen, and will continue to see, benefits from their use. For instance, ML algorithms have found applications in providing accurate SN of PET images, which aids in the classification process of AD. ML algorithms have also aided in the processing stages of PET imaging, primarily with respect to image reconstruction and AC, thereby obtaining a higher resolution and a more accurate PET image that further aids the physician in making informed medical decisions based on the PET data. Furthermore, ML algorithms have been utilized to help produce satisfactory PET images while reducing the quantity of ionizing radiation that the patient experiences, either by predicting PET images with the spatial resolution of a standard dose of radiotracer while using the data from a lowered dose or by producing accurate AC for PET or PET/MR without the need for an accompanying CT scan. Predictive and classification software, for instance, in the early diagnosis of AD or in cancer stratification and prognosis, will provide fast decision support for physicians diagnosing patients and selecting the most effective treatment for individuals. There are currently 4 oncologic clinical trials that are applying ML to PET images for characterization or predicting response to treatment (ClinicalTrials.gov identifiers NCT00330109, NCT03594760, NCT03574454, NCT03517306), suggesting this to be a growing area of research. As with most new technologies, the successful implementation of ML algorithms also comes with limitations: namely, the utility of the ML algorithm is restricted by the scope and breadth of the data set with which it was provided for training. Hence, although the availability of publicly available datasets such as ADNI has promoted the development of many of the ML algorithms discussed in this article, it is important that such algorithms be cross-validated with other datasets to ensure general applicability prior to their introduction to a wider population.7 With the potential to support the patient diagnosis process—thereby improving personalized treatment regimens—as well as its beneficial impact on PET imaging protocols, the current applications of ML algorithms in PET, as surveyed by this article, provide a platform for an exciting frontier with the potential to hugely impact the outcomes of patients with cancer and/or neurodegenerative diseases.
It is envisioned that ML algorithms will complement the innovations that will drive the future of molecular imaging, including miniaturization and mobile technologies such as wearable helmet PET scanners78 and mini-cyclotrons (ABT Molecular Imaging Inc; USA). Many of these inventions would be further promoted by the practical utility of ultralow tracer dosages, enabled by the use of ML algorithms. Such technologies will likely make PET more accessible to a larger subset of the general population, thereby producing even larger datasets and necessitating the need for ML algorithms to perform analysis. Such examples are just some of the potential applications of ML algorithms as the future of healthcare moves toward precision health.79
Footnotes
Authors’ Note: Ian R. Duffy and Amanda J. Boyle are co-first authors.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: N.V. thanks National Institute on Ageing of the NIH (R01AG054473 and R01AG052414), the Azrieli Foundation, and the Canada Research Chairs Program for support.
ORCID iD: Neil Vasdev  https://orcid.org/0000-0002-2087-5125
https://orcid.org/0000-0002-2087-5125
References
- 1. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi:10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acharya UR, Hagiwara Y, Sudarshan VK, Chan WY, Ng KH. Towards precision medicine: from quantitative imaging to radiomics. J Zhejiang Univ Sci B. 2018;19(1):6–24. doi:10.1631/jzus.B1700260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. Deep learning with convolutional neural network in radiology. Jpn J Radiol. 2018;36(4):257–272. doi:10.1007/s11604-018-0726-3. [DOI] [PubMed] [Google Scholar]
- 4. Kohli M, Prevedello LM, Filice RW, Geis JR. Implementing machine learning in radiology practice and research. AJR Am J Roentgenol. 2017;208(4):754–760. doi:10.2214/ajr.16.17224. [DOI] [PubMed] [Google Scholar]
- 5. Vallières M, Zwanenburg A, Badic B, Cheze Le Rest C, Visvikis D, Hatt M. Responsible radiomics research for faster clinical translation. J Nucl Med. 2018;59(2):189–193. doi:10.2967/jnumed.117.200501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thrall JH, Li X, Li Q. et al. Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J Am Coll Radiol. 2018;15(3 pt B):504–508. doi:10.1016/j.jacr.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 7. Smith SM, Nichols TE. Statistical challenges in “big data” human neuroimaging. Neuron. 2018;97(2):263–268. doi:10.1016/j.neuron.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 8. Liu X, Chen K, Wu T, Weidman D, Lure F, Li J. Use of multimodality imaging and artificial intelligence for diagnosis and prognosis of early stages of Alzheimer’s disease. Transl Res. 2018;194:56–67. doi:10.1016/j.trsl.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tseng HH, Wei L, Cui S, Luo Y, Ten Haken RK, El Naqa I. Machine learning and imaging informatics in oncology. Oncology. 2018:1–19. doi:10.1159/000493575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. As this manuscript was being reviewed, a separate review article detailing the applications of ML algorithms in PET/MR and PET/CT imaging was made available. See: Zaharchuk G. Next generation research applications for hybrid PET/MR and PET/CT imaging using deep learning. Eur J Nucl Med Mol Imaging. 2019; doi:10.1007/s00259-019-04374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uribe CF, Mathotaarachchi S, Gaudet V. et al. Machine learning in nuclear medicine: part 1-introduction. J Nucl Med 2019;60(4):451–458. doi:10.2967/jnumed.118.223495. [DOI] [PubMed] [Google Scholar]
- 12. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi:10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 13. Petersen RC, Aisen PS, Beckett LA. et al. Alzheimer’s disease neuroimaging initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–209. doi:10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chauvie S, Bertone E, Bergesio F, Terulla A, Botto D, Cerello P. Automatic liver detection and standardised uptake value evaluation in whole-body positron emission tomography/computed tomography scans. Comput Meth Programs Biomed. 2018;156:47–52. doi:10.1016/j.cmpb.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 15. Clark K, Vendt B, Smith K. et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. doi:10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bakr S, Gevaert O, Echegaray S. et al. A radiogenomic dataset of non-small cell lung cancer. Sci Data. 2018;5:180202 doi:10.1038/sdata.2018.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martinez-Moller A, Souvatzoglou M, Delso G. et al. Tissue classification as a potential approach for attenuation correction in whole-body PET/MRI: evaluation with PET/CT data. J Nucl Med. 2009;50(4):520–526. doi:10.2967/jnumed.108.054726. [DOI] [PubMed] [Google Scholar]
- 18. Spick C, Herrmann K, Czernin J. 18F-FDG PET/CT and PET/MRI perform equally well in cancer: evidence from studies on more than 2,300 patients. J Nucl Med. 2016;57(3):420–430. doi:10.2967/jnumed.115.158808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Samarin A, Burger C, Wollenweber SD. et al. PET/MR imaging of bone lesions—implications for PET quantification from imperfect attenuation correction. Eur J Nucl Med Mol Imaging. 2012;39(7):1154–1160. doi:10.1007/s00259-012-2113-0. [DOI] [PubMed] [Google Scholar]
- 20. Yang J, Park D, Gullberg GT, Seo Y. Joint correction of attenuationg and scatter in image space using deep convolutional neural networks for dedicated brain 18F-FDG PET. Phys Med Biol. 2019;64(7):075019. doi:10.1088/1361-6560/ab0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB. A deep learning approach for 18F-FDG PET attenuation correction. EJNMMI Phys. 2018;5(1):24 doi:10.1186/s40658-018-0225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ladefoged CN, Marner L, Hindsholm A, Law I, Højgaard L, Andersen FL. Deep learning based attenuation correction of PET/MRI in pediatric brain tumor patients: evaluation in a clinical setting. Front Neurosci. 2019;12:1005 doi:10.3389/fnins.2018.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ladefoged CN, Benoit D, Law I. et al. Region specific optimization of continuous linear attenuation coefficients based on UTE (RESOLUTE): application to PET/MR brain imaging. Phys Med Biol. 2015;60(20):8047–8065. doi:10.1088/0031-9155/60/20/8047. [DOI] [PubMed] [Google Scholar]
- 24. Torrado-Carvajal A, Vera-Olmos J, Izquierdo-Garcia D. et al. Dixon-VIBE deep learning (DIVIDE) pseudo-CT synthesis for pelvis PET/MR attenuation correction. J Nucl Med. 2018;60(3):429–435. doi:10.2967/jnumed.118.209288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hwang D, Kang SK, Kim KY. et al. Generation of PET attenuation map for whole-body time-of-flight 18F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med. 2019;60(8):1183–1189. doi:10.2967/jnumed.118.219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bradshaw TJ, Zhao G, Jang H, Liu F, McMillan AB. Feasibility of deep learning-based PET/MR attenuation correction in the pelvis using only diagnostic MR images. Tomography. 2018;4(3):138–147. doi:10.18383/j.tom.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Häggström I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: a deep encoder–decoder network for directly solving the PET image reconstruction inverse problem. Med Image Anal. 2019;54:253–262. doi:10.1016/j.media.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang B, Ying L, Tang J. Artificial neural network enhanced bayesian PET image reconstruction. IEEE Trans Med Imaging. 2018;37(6):1297–1309. doi:10.1109/tmi.2018.2803681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang J, Yang B, Wang Y, Ying L. Sparsity-constrained PET image reconstruction with learned dictionaries. Phys Med Biol. 2016;61(17):6347–6368. doi:10.1088/0031-9155/61/17/6347. [DOI] [PubMed] [Google Scholar]
- 30. Liu C-C, Qi J. Higher SNR PET image prediction using a deep learning model and MRI image. Phys Med Biol. 2019;64(11):115004. doi:10.1088/1361-6560/ab0dc0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kang SK, Seo S, Shin SA. et al. Adaptive template generation for amyloid PET using a deep learning approach. Hum Brain Mapp. 2018;39:3769–3778. doi:10.1002/hbm.24210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen Y, Li S, Brown C. et al. Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet Genomics. 2009;19(7):497–504. doi:10.1097/FPC.0b013e32832cc7e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen KT, Gong E, Macruz FBdC. et al. Ultra–low-dose 18F-florbetaben amyloid PET imaging using deep learning with multi-contrast MRI inputs. Radiology. 2019;290(3):649–656. doi:10.1148/radiol.2018180940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang B, Fontaine K, Carson R, Tang J. Brain PET dose reduction using a shallow artificial neural network. J Nucl Med. 2018;59:99a (Abstract). [Google Scholar]
- 35. Xiang L, Qiao Y, Nie D. et al. Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing. 2017;267:406–416. doi:10.1016/j.neucom.2017.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaplan S, Zhu Y-M. Full-dose PET image estimation from low-dose PET image using deep learning: a pilot study. J Digit Imaging. 2018. doi:10.1007/s10278-018-0150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwyzer M, Ferraro DA, Muehlematter UJ. et al. Automated detection of lung cancer at ultralow dose PET/CT by deep neural networks – initial results. Lung Cancer. 2018;126:170–173. doi:10.1016/j.lungcan.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 38. Rice L, Bisdas S. The diagnostic value of FDG and amyloid PET in Alzheimer’s disease—a systematic review. Eur J Radiol. 2017;94:16–24. doi:10.1016/j.ejrad.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 39. Jagust W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat Rev Neurosci. 2018;19(11):687–700. doi:10.1038/s41583-018-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smailagic N, Vacante M, Hyde C, Martin S, Ukoumunne O, Sachpekidis C. 18F-FDG PET for the early diagnosis of Alzheimer’s disease dementia and other dementias in people with mild cognitive impairment (MCI). Cochrane Database Syst Rev. 2015. doi:10.1002/14651858.CD010632.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ding Y, Sohn JH, Kawczynski MG. et al. A deep learning model to predict a diagnosis of Alzheimer disease by using 18F-FDG PET of the brain. Radiology. 2019;290(2):456–464. doi:10.1148/radiol.2018180958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu D, Popuri K, Ding GW, Balachandar R, Beg MF; Alzheimer’s Disease Neuroimaging Initiative. Multiscale deep neural network based analysis of FDG-PET images for the early diagnosis of Alzheimer’s disease. Med Image Anal. 2018;46:26–34. doi:10.1016/j.media.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 43. Lu D, Popuri K, Ding GW, Balachandar R, Beg MF; Alzheimer’s Disease Neuroimaging Initiative. Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s disease using structural MR and FDG-PET images. Sci Rep. 2018;8(1):5697 doi:10.1038/s41598-018-22871-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Choi H, Jin KH; Alzheimer’s Disease Neuroimaging Initiative. Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav Brain Res. 2018;344:103–109. doi:10.1016/j.bbr.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 45. Gamberger D, Lavrač N, Srivatsa S, Tanzi RE, Doraiswamy PM. Identification of clusters of rapid and slow decliners among subjects at risk for Alzheimer’s disease. Sci Rep. 2017;7(1):6763 doi:10.1038/s41598-017-06624-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu M, Cheng D, Yan W; Alzheimer’s Disease Neuroimaging Initiative. Classification of Alzheimer’s disease by combination of convolutional and recurrent neural networks using FDG-PET images. Front Neuroinform. 2018;12:35 doi:10.3389/fninf.2018.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu M, Cheng D, Wang K, Wang Y; Alzheimer’s Disease Neuroimaging Initiative. Multi-modality cascaded convolutional neural networks for Alzheimer’s disease diagnosis. Neuroinformatics. 2018;16(3-4):295–308. doi:10.1007/s12021-018-9370-4. [DOI] [PubMed] [Google Scholar]
- 48. Zhou T, Thung K-H, Zhu X, Shen D. Effective feature learning and fusion of multimodality data using stage-wise deep neural network for dementia diagnosis. Hum Brain Mapp. 2019; 40(3):1001–1016.doi:10.1002/hbm.24428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Whittington A, Sharp DJ, Gunn RN; Alzheimer’s Disease Neuroimaging Initiative. Spatiotemporal distribution of β-amyloid in Alzheimer disease is the result of heterogeneous regional carrying capacities. J Nucl Med. 2018;59(5):822–827. doi:10.2967/jnumed.117.194720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Whittington A, Gunn RN; Alzheimer’s Disease Neuroimaging Initiative. Amyloid load: a more sensitive biomarker for amyloid imaging. J Nucl Med. 2018;60(4):536–540. doi:10.2967/jnumed.118.210518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Whittington A, Seibyl J, Hesterman J, Gunn R. TauIQ—an algorithm to quantify global and local tau accumulation. Poster Session Presented at the 13th Human Amyloid Imaging Conference; January 2019; Miami, FL. [Google Scholar]
- 52. Chauvie S, Biggi A, Stancu A. et al. WIDEN: a tool for medical image management in multicenter clinical trials. Clin Trials. 2014;11(3):355–361. doi:10.1177/1740774514525690. [DOI] [PubMed] [Google Scholar]
- 53. Runa F, Hamalian S, Meade K, Shisgal P, Gray PC, Kelber JA. Tumor microenvironment heterogeneity: challenges and opportunities. Curr Mol Biol Rep. 2017;3(4):218–229. doi:10.1007/s40610-017-0073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cook GJR, Azad G, Owczarczyk K, Siddique M, Goh V. Challenges and promises of PET radiomics. Int J Radiat Oncol Biol Phys. 2018;102(4):1083–1089. doi:10.1016/j.ijrobp.2017.12.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–354. doi:10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 56. Chen SW, Shen WC, Hsieh TC. et al. Textural features of cervical cancers on FDG-PET/CT associate with survival and local relapse in patients treated with definitive chemoradiotherapy. Sci Rep. 2018;8(1):11859 doi:10.1038/s41598-018-30336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yip SS, Coroller TP, Sanford NN, Mamon H, Aerts HJ, Berbeco RI. Relationship between the temporal changes in positron-emission-tomography-imaging-based textural features and pathologic response and survival in esophageal cancer patients. Front Oncol. 2016;6:72 doi:10.3389/fonc.2016.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cheng NM, Fang YH, Lee LY. et al. Zone-size nonuniformity of 18F-FDG PET regional textural features predicts survival in patients with oropharyngeal cancer. Eur J Nucl Med Mol Imaging. 2015;42(3):419–428. doi:10.1007/s00259-014-2933-1. [DOI] [PubMed] [Google Scholar]
- 59. Xiong J, Yu W, Ma J. et al. The role of PET-based radiomic features in predicting local control of esophageal cancer treated with concurrent chemoradiotherapy. Sci Rep. 2018;8:9902 doi:10.1038/s41598-018-28243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mattonen SA, Davidzon GA, Bakr S. et al. [18F]FDG positron emission tomography (PET) tumor and penumbra imaging features predict recurrence in non–small cell lung cancer. Tomography. 2019;5(1):145–153. doi:10.18383/j.tom.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. 2010;49(7):1012–1016. doi:10.3109/0284186x.2010.498437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Leijenaar RT, Nalbantov G, Carvalho S. et al. The effect of SUV discretization in quantitative FDG-PET radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075 doi:10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Berthon B, Marshall C, Evans M, Spezi E. ATLAAS: an automatic decision tree-based learning algorithm for advanced image segmentation in positron emission tomography. Phys Med Biol. 2016;61(13):4855–4869. doi:10.1088/0031-9155/61/13/4855. [DOI] [PubMed] [Google Scholar]
- 64. Berthon B, Evans M, Marshall C. et al. Head and neck target delineation using a novel PET automatic segmentation algorithm. Radiother Oncol. 2017;122(2):242–247. doi:10.1016/j.radonc.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 65. Huang B, Chen Z, Wu PM. et al. Fully automated delineation of gross tumor volume for head and neck cancer on PET-CT using deep learning: a dual-center study. Contrast Media Mol Imaging. 2018:8923028. doi:10.1155/2018/8923028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Blanc-Durand P, Van Der Gucht A, Schaefer N, Itti E, Prior JO. Automatic lesion detection and segmentation of 18F-FET PET in gliomas: a full 3D U-Net convolutional neural network study. PLoS One. 2018;13(4):e0195798 doi:10.1371/journal.pone.0195798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dunet V, Pomoni A, Hottinger A, Nicod-Lalonde M, Prior JO. Performance of 18F-FET versus 18F-FDG-PET for the diagnosis and grading of brain tumors: systematic review and meta-analysis. Neuro Oncol. 2016;18(3):426–434. doi:10.1093/neuonc/nov148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Blanc-Durand P, Van Der Gucht A, Verger A. et al. Voxel-based 18F-FET PET segmentation and automatic clustering of tumor voxels: a significant association with IDH1 mutation status and survival in patients with gliomas. PLoS One. 2018;13(6):e0199379 doi:10.1371/journal.pone.0199379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang H, Zhou Z, Li Y. et al. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res. 2017;7(1):11 doi:10.1186/s13550-017-0260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. De Bernardi E, Buda A, Guerra L. et al. Radiomics of the primary tumour as a tool to improve 18F-FDG-PET sensitivity in detecting nodal metastases in endometrial cancer. EJNMMI Res. 2018;8(1):86 doi:10.1186/s13550-018-0441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen L, Zhou Z, Sher D. et al. Combining many-objective radiomics and 3D convolutional neural network through evidential reasoning to predict lymph node metastasis in head and neck cancer. Phys Med Biol. 2019;64(7):075011 doi:10.1088/1361-6560/ab083a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Toney LK, Vesselle HJ. Neural networks for nodal staging of non-small cell lung cancer with FDG PET and CT: importance of combining uptake values and sizes of nodes and primary tumor. Radiology. 2014;270(1):91–98. doi:10.1148/radiol.13122427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Milgrom SA, Elhalawani H, Lee J. et al. A PET radiomics model to predict refractory mediastinal hodgkin's lymphoma. Sci Rep. 2019;9(1):1322 doi:10.1038/s41598-018-37197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ypsilantis PP, Siddique M, Sohn HM. et al. Predicting response to neoadjuvant chemotherapy with PET imaging using convolutional neural networks. PLoS One. 2015;10(9):e0137036 doi:10.1371/journal.pone.0137036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Riyahi S, Choi W, Liu CJ. et al. Quantifying local tumor morphological changes with Jacobian map for prediction of pathologic tumor response to chemo-radiotherapy in locally advanced esophageal cancer. Phys Med Biol. 2018;63(14):145020 doi:10.1088/1361-6560/aacd22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kirienko M, Sollini M, Silvestri G. et al. Convolutional neural networks promising in lung cancer T-parameter assessment on baseline FDG-PET/CT. Contrast Media Mol Imaging. 2018;2018:1382309 doi:10.1155/2018/1382309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tsujikawa T, Rahman T, Yamamoto M. et al. 18F-FDG PET radiomics approaches: comparing and clustering features in cervical cancer. Ann Nucl Med. 2017;31(9):678–685. doi:10.1007/s12149-017-1199-7. [DOI] [PubMed] [Google Scholar]
- 78. Melroy S, Bauer C, McHugh M. et al. Development and design of next-generation head-mounted ambulatory microdose positron-emission tomography (AM-PET) system. Sensors. 2017;17(5):1164. doi:10.3390/s17051164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gambhir SS, Ge TJ, Vermesh O, Spitler R. Toward achieving precision health. Sci Transl Med. 2018;10(430):eaao3612 doi:10.1126/scitranslmed.aao3612. [DOI] [PMC free article] [PubMed] [Google Scholar]