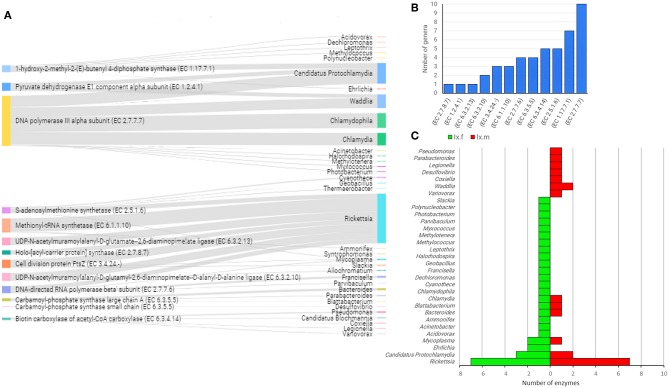
Figure 8.
Linkage of enzyme-encoding genes with bacterial genera harboring them into the gut microbiome of both IO and IP ticks. Only enzyme-encoding genes with significant differences in relative abundance (q < 0.05) between male and female ticks and KEGG annotation were included in this analysis (A). Sankey graph showing the connectivity between functional genes (left) and bacterial genera (right) wherein those were annotated. The edges link genes and source (bacterial genera). The node size is proportional to the number of hits (number of sequences containing the annotation), and consequently the edges size is proportional to the hit's distribution among bacterial genera. (B) Bar plot indicating the number of bacterial genera harboring every enzyme-encoding gene. (C) Mirror bar plot showing the distribution of enzyme-encoding genes across the bacterial genera in female (Ix.f) and male (Ix.m) Ixodes ticks (data on both ticks species, IO and IP, was merged). A detailed list of enzymes and bacterial source can be found in Supplementary Table 3.