Figure 3. Centromeric nucleosome interactions with non-histones, CENP-C and CENP-N.
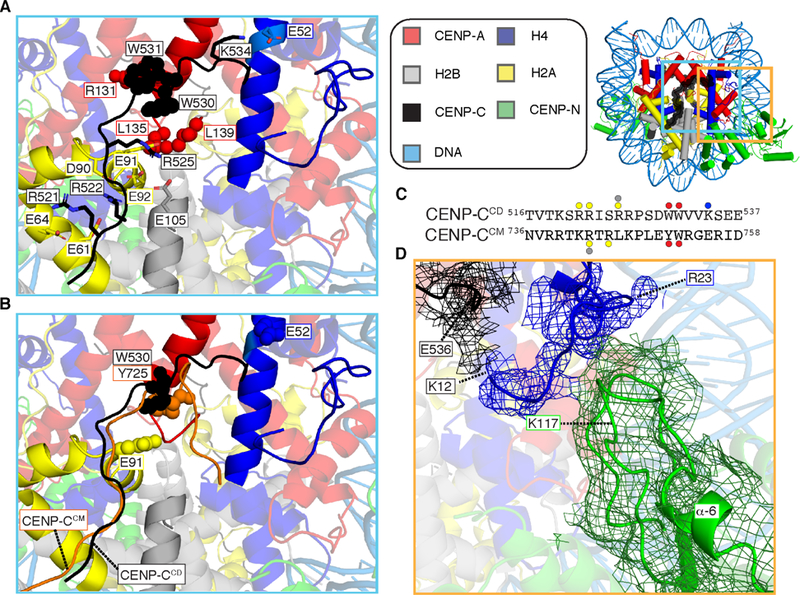
(A) CENP-Ca.a518−537 bonds with all four histone subunits of the centromeric nucleosome involve extensive electrostatic (sticks) and hydrophobic (spheres) interactions (See also Figure S4E) (B) CENP-CCM (orange) from PDB# 4X23 is aligned with the CCNC structure. The additional electrostatic interactions made by CENP-CCD with sites on the histone octamer (H2AE91 and H4E52) are shown in space fill, as are CENP-CW530 and CENP-CY725 at the site of contact with CENP-A or histone H3 C-termini, in CENP-CCD and CENP-CCM, respectively. (C) Protein sequence alignment of human CENP-CCD and CENP-CCM. The sites of contact with histone components are highlighted with circles color coded to match the histone subunits. (D) Cryo-EM density for the histone tail of H4 (mesh overlaying ribbon model) extends to H4K12, with contacts between H4a.a.12−20 with the C-terminal portion of CENP-CCD on one side and two nearby loops of CENP-NNT on the other. Density here was assigned to the H4 tail prior to B-factor correction, and H4R23 is labeled because its side-chain density in our map provided a landmark with which to orient main chain density N-terminal to it. Local refinement (see EMDB #9252 and see also Figure S5A) yielded the shown density map.
