Figure 4. DNA sequence contributes to the structure and faithful assembly of the CCNC.
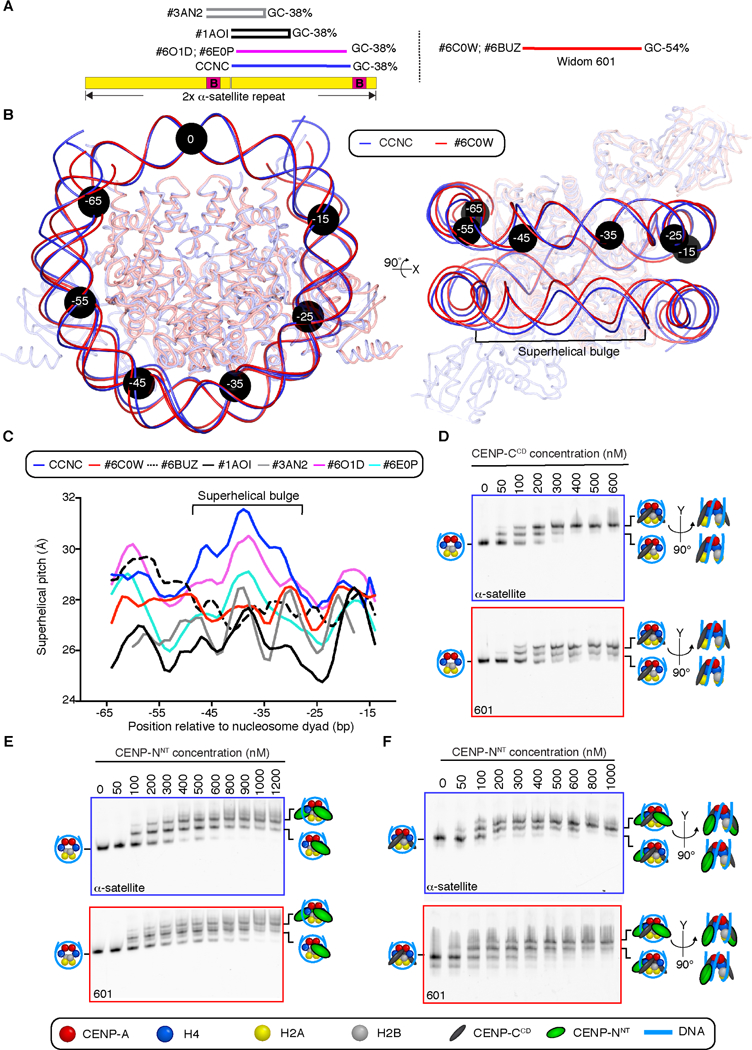
(A) Position of on a tandem copy of the 171 bp human α-satellite DNA sequence of palindromic sequences used in prior nucleosome structure studies [29,30], the natural sequence used in this study of the CCNC, and the unnatural “Widom 601” sequence that has emerged as the most common sequence for nucleosome structural studies. (B) Alignment using one CENP-A and histone H4 dimer of CCNC structure with PDB# 6C0W (CENP-A nucleosome wrapped with 601 DNA and bound by one copy of CENP-NNT; [21]. Black circles denote the indicated number of bp from the nucleosome dyad. Note that the DNA used for both structural studies is 147 bp of DNA, although the terminal 4 bp are not resolved in PDB# 6C0W (see also Figure S6). (C) Local superhelical pitch measurements comparing the interphase CCNC with the indicated related structures in the PDB. (D) Binding of CENP-CCD to CENP-A nucleosomes wrapped with either α-satellite or 601 DNA. (E) Binding of CENP-NNT to CENP-A nucleosomes wrapped with either α-satellite or 601 DNA. (F) Binding of CENP-NNT to CENP-A nucleosomes wrapped with either α-satellite or 601 DNA and bound by CENP-CCD. Binding assays in D-F are representative example of three independent experiments.
