Abstract
Flying insects engage in spectacular high-speed pursuit of targets, requiring visual discrimination of moving objects against cluttered backgrounds. As a first step toward understanding the neural basis for this complex task, we used computational modeling of insect small target motion detector (STMD) neurons to predict responses to features within natural scenes and then compared this with responses recorded from an identified STMD neuron in the dragonfly brain (Hemicordulia tau). A surprising model prediction confirmed by our electrophysiological recordings is that even heavily cluttered scenes contain very few features that excite these neurons, due largely to their exquisite tuning for small features. We also show that very subtle manipulations of the image cause dramatic changes in the response of this neuron, because of the complex inhibitory and facilitatory interactions within the receptive field.
Introduction
Insects engage in high-speed aerobatics while visually detecting and tracking prey or conspecifics (Collett and Land, 1978; Olberg et al., 2000). This requires visualizing the moving object against an often highly cluttered background scene, which itself may be in rapid motion due to the animal's rotational and translational body movement. This ability is remarkable considering insects have a visual acuity less than one-sixtieth that of human vision (Horridge, 1978).
Neurons likely to underlie such behavior, with exquisite tuning for small moving targets, have been recorded from the ventral nerve cord in dragonflies (Olberg, 1981) and the optic lobes in dragonflies and dipteran flies (Collett and King, 1975; O'Carroll, 1993; Nordström et al., 2006; Barnett et al., 2007; Geurten et al., 2007). These small target motion detector (STMD) neurons respond selectively to small objects moving within their excitatory receptive field regardless of the target location (i.e., display position invariance in object-size tuning). Many properties of these neurons resemble those of hypercomplex cells found in the mammalian cortex (O'Carroll, 1993). A subset respond robustly to a target against a cluttered, moving background, even when their respective velocities are matched (Nordström et al., 2006; Nordström and O'Carroll, 2009).
CSTMD1 (Centrifugal Small Target Motion Detector 1), a recently identified giant STMD neuron from the dragonfly Hemicordulia tau, has become a promising model system for in-depth analysis of neural mechanisms underlying target discrimination (Geurten et al., 2007). CSTMD1 has inputs in the lateral midbrain and an axon that traverses the brain. It makes two outputs in the contralateral brain. The first is an extensive arborization across the entire contralateral lobula (third optic ganglion). Interestingly, the second output region, in the contralateral midbrain, is coincident with the input region of CSTMD1's mirror symmetric counterpart. This raises the intriguing possibility of communication between the two neurons to mediate a simple form of visual attention. Bolzon et al. (2009) showed that the simultaneous presence of a second target in the contralateral hemisphere completely suppresses the response to a target in the excitatory receptive field. In an additional experiment, they presented a second target within this excitatory region, revealing a large and diffuse inhibitory surround. In a more recent investigation, Nordström et al. (2011) examined a form of spatial facilitation for targets moving along long trajectories within the receptive field. Hence, CSTMD1 is a higher-order neuron, useful for studying both the underlying mechanisms for target tuning and more complex higher-order interactions among local motion detectors that may shape responses to multiple targets or features.
To date, experimental analysis of STMDs has been limited to very simple visual stimuli. In contrast, natural scenes contain complex textures and a rich mixture of spatial frequencies that might match the target tuning of an STMD neuron (Field, 1987). The question then arises as to which features within a natural scene might evoke a response? To test this, we presented three versions of natural scenes to dragonflies while recording from CSTMD1. The first version was unaltered and in the second we embedded an optimal-size target. The third was digitally manipulated by a computational model for small target motion detection to remove features predicted to excite CSTMD1.
Materials and Methods
Computational modeling and image processing.
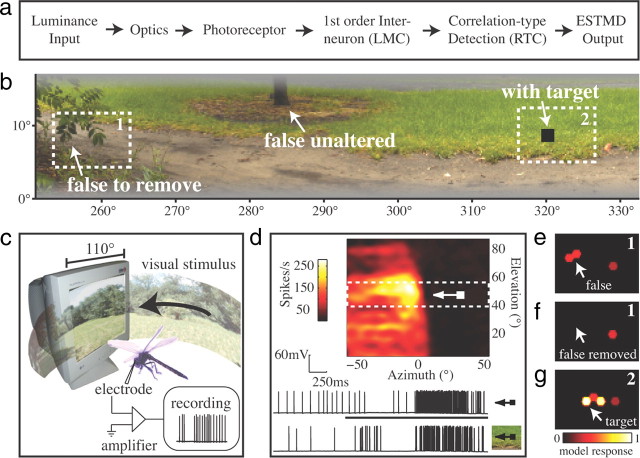
A model for elementary (local) small target motion detectors (ESTMDs) was written in MATLAB (Fig. 1a). Full details of the model are given in Wiederman et al. (2008, 2010), but briefly, the model encapsulates insect optics (blurring and hexagonal sampling) and early visual processing [dynamic bandpass filtering of the photoreceptors and large monopolar cells (LMC)]. Size selectivity is conferred by a bio-inspired stage based on physiological characteristics of rectifying transient cells (RTC), a class of ON/OFF neuron recorded from the insect medulla (Osorio, 1991; Wiederman et al., 2008). Half-wave rectification separates the contrast signal into separate ON and OFF channels, further processed via fast temporal adaptation and strong surround antagonism. These are then recombined by multiplication of the delayed OFF signal with the undelayed ON signal, resulting in a spatiotemporal filter matched to both the contrast boundaries and limited spatial extent of a passing (dark) target. The model also includes a further elaboration, a potent inhibitory surround, as evidenced by recent electrophysiological recordings (Bolzon et al., 2009). To identify features likely to elicit a response from STMD neurons, we animated panoramic scenes horizontally past the ESTMD model inputs (for detailed methods, see Wiederman et al., 2010). We then created two additional versions of the image.
Figure 1.
a, An overview of the ESTMD model. b, A section from a natural scene (bushes) illustrating false-positive features that were either left intact or removed by digital manipulation, and an optimal black target (1.6° × 1.6°) added to one version of the image (see Materials and Methods). c, Illustration of the experimental recording setup, with panoramic textures animated on a 200 Hz CRT display. d, Receptive field of CSTMD1, measured by drifting a target (0.8° × 0.8°) across the CRT monitor at 21 elevations. The upper raw data (inset) illustrates the response to the target alone through the receptive field center (black stimulus bar denotes duration of the target on the monitor screen). The lower raw data show a typical response to the target when embedded in the bushes scene (of an equivalent duration). Note the suppression of spike rate as a target traverses the visual field of the contralateral eye. e–g, ESTMD model responses to the rectangular regions (1 and 2 in b) highlighted in three variants of the image. e, Region 1 of the original image with a group of leaves that form a false-positive feature. f, Region 1 after digital manipulation to remove the group of leaves. g, Region 2 in the image with the target included.
In the first additional image (image plus target), we embedded an optimal-size target (1.6° × 1.6°) on a low-density part of the background texture (Fig. 1b). The ESTMD model response to the image plus target is shown in Figure 1g. For clarity, only the model response to region 2 (Fig. 1b) is displayed. The target produced the strongest response (white), which is smeared to the right due to the temporal filtering inherent in the model. Note that the model response strengthens and weakens as the target passes horizontally across the facets because their hexagonal arrangement results in slightly different vertical position relative to the target trajectory.
To create the second additional image (image minus false positive), features predicted by the ESTMD model as potential false targets were removed using the Healing Brush tool (Adobe Photoshop CS3), which is commonly used by artists to remove blemishes from photographs. This process replaced the feature with a background texture sampled from nearby locations. The model responses to region 1 (Fig. 1b) both before and after the digital manipulation (with and without the false positive) are shown in Figure 1, e and f, respectively.
Electrophysiology.
Seven wild-caught, male dragonflies (Hemicordulia tau) were immobilized with a beeswax and rosin (1:1) mixture and the head was tilted forward to access the posterior surface. A hole was cut above the lobula where we recorded intracellularly with aluminum silicate micropipettes pulled on a Sutter Instruments P-97 puller and backfilled with KCl (2 m). The electrode tip resistance was typically ∼110 MΩ.
Stimulus presentation.
The dragonfly was positioned facing the center of a high refresh rate (200 Hz) CRT monitor upon which visual stimuli were presented using VisionEgg software (www.visionegg.org). The display subtended ∼110° × 80° of the animal's visual field of view, with a resolution of 640 × 480 pixels. Data were digitized using LabView software at 5 kHz using a 16-bit A/D converter (National Instruments) and analyzed off-line with Matlab (Mathworks).
CSTMD1 was identified by its distinctive receptive field, mapped with a drifting target stimulus (Fig. 1d) and its characteristic physiology, with large biphasic action potentials (unusual for an STMD neuron) and somewhat regular spontaneous activity. We also drifted targets of varying heights through the receptive field center, with the characteristic tuning curve and peak firing rate to optimal targets providing additional verification of the identification of CSTMD1 (data not shown). Following identification, all three versions of the image were animated in front of the dragonfly (Fig. 1c).
The 360° panoramic image moved through the neuron's receptive field at the preferred velocity (45°/s) with 110° horizontal extent of the image viewable on the monitor at any moment. The images were carefully centered within the hotspot of the receptive field, although small variations in this positioning and CSTMD1's receptive field structure between animals might account for some of the variation in CSTMD1 responses observed.
Results
Figure 2 shows CSTMD1 responses to repeated presentations of the three versions of a natural scene obtained where dragonflies were active (bushes) (Fig. 2a–d) and a second from a manmade environment (car park) (Fig. 2e–h). For clarity, only the versions containing the artificially embedded target are shown. The electrophysiological responses are represented as averaged spike histograms (Fig. 2, b–d, f–h, blue lines) and raster plots for individual neurons (seven neurons in seven dragonflies) (Fig. 2, b–d, f–h). Because both the scenes and the receptive field of CSTMD1 are large, it is difficult to be certain which features presented within the receptive field elicited observed responses. We therefore aligned recorded responses with the part of the stimulus image that was present within the receptive field hotspot (Fig. 1d) at the corresponding time—a region that would contribute disproportionately to the overall response. Furthermore, although the original images used in modeling were 72° in vertical extent, we limited the display to a 14° strip centered on the most prominent false-positive feature, ensuring that these features passed through the receptive field center.
Figure 2.
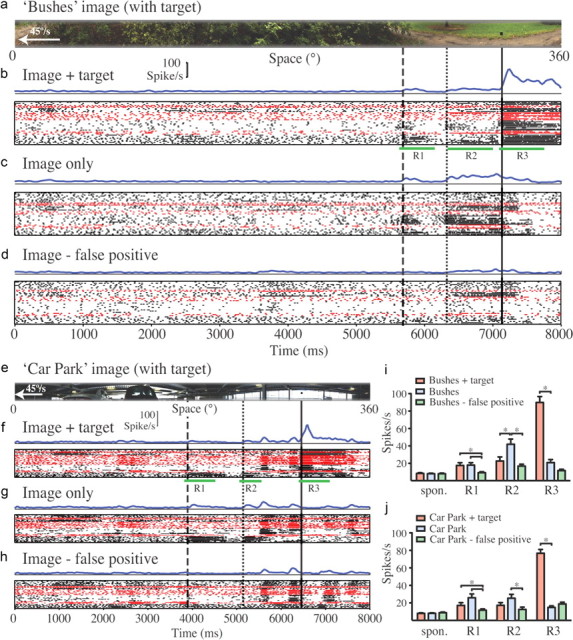
Electrophysiological responses of CSTMD1 to image strips moved at 45°/s from right to left across the frontal visual field. All responses are represented by average spike histograms (blue lines), raster plots where dots represent individual spikes recorded, and rows representing presentations of the same stimulus. Individual CSTMD1 neuron recordings from seven different animals' cells are distinguished by alternating black and red symbols. a, The strip from the bushes natural scene presented in physiological recordings, shown in alignment with recorded responses (see Results). b, Neural responses to the scene with the inclusion of an optimal target. R1, R2, and R3 denote peristimulus periods used for analysis of responses as three features identified by vertical lines passing through the receptive field center. The dashed line and R1 are aligned with the false-positive leaf feature (Fig. 1b,c). The dotted line (R2) is aligned with the base of a tree trunk (Fig. 1b) that was not altered during experiments. The solid vertical line (R3) is aligned with the embedded target feature. c, Responses to the original image. Some neurons respond to the false-positive feature (R1) and the tree base (R2). d, Responses to the image after digital manipulation to remove the false-positive feature (dashed line and R1). e–h, Similar analysis for the car park scene from a manmade environment. Note more transient responses to the target (R3) in f compared with same feature in the bushes image (b). Some neurons respond to the false-positive feature (at R1 in f and g) and to column features around R2 (dotted line). i, j, Summary analysis for each of the three image versions and both scenes, averaged across all seven neurons (between 37 and 51 repetitions for each image) and within the windows identified by R1–R3. *p < 0.05 (two-tailed t test with Bonferroni multiple comparisons, following one-way ANOVA) for comparison of response changes within each subregion (R1–R3) between images.
CSTMD1 responded only weakly (near spontaneous levels) to background clutter within the bushes image until the embedded target entered the receptive field at ∼7100 ms (Fig. 2b, solid line). A sustained response endured for 800 ms as the target drifted through the excitatory receptive field (36° wide). We also observed responses in some neurons to a small group of leaves identified by the model as the strongest false-positive feature (Fig. 2b,c, dashed line). When these were digitally removed, the corresponding responses were significantly reduced to near spontaneous levels (Fig. 2d; i, R1). Interestingly, this same manipulation also significantly reduced the response to an unaltered feature, the base of a small tree (Fig. 2c, dotted line; i, R2). This feature (Fig. 1b) only comes close to matching the preferred target size of CSTMD1 as an artifactual consequence of our limiting the vertical extent of the image as displayed to CSTMD1.
Our recordings highlight the lack of response of these neurons to background texture within heavily cluttered scenes while retaining the ability to respond robustly to optimal-size features. In manmade environments, we might expect the preponderance of hard vertical boundaries to be more challenging. Nevertheless CSTMD1 also discriminates the target from background in the car park image (Fig. 2f, solid line). Interestingly, the response was more transient than the response to the same target embedded in the bushes image (compare Fig. 2, b and f). Again, there were weak responses to a false-positive feature predicted by the ESTMD model (Fig. 2f,g, dashed lines) in some recorded neurons. The digital removal of this feature significantly reduced the corresponding neural responses (Fig. 2h; j, R1). There were weak responses to the vertical building columns in all image versions (t = 5600 ms, 6200 ms), also vertically truncated by our selection of the narrow image strip as displayed to CTSMD1. Interestingly, as in the bushes image, the response to one of these features (Fig. 2f,g, dotted lines) was also significantly reduced by removal of the false-positive feature (Fig. 2h; j, R2).
Although our ESTMD model was able to predict a small number of false-positive background features within natural scenes that would be expected to stimulate STMD neurons, our measured responses were typically weak and notably variable between recordings. Coefficients of variation (CV) are more than double (CV = 1.04 and 1.19 for R1 in the bushes and car park scenes, respectively) compared with the responses to the artificially embedded target (CV = 0.49 and 0.40 for R3 in the image versions with the embedded targets) (Fig. 2b,f).
Discussion
Despite the intuitive concept that natural, or even manmade, environments may contain numerous features matching the size preference of STMD neurons, our modeling suggests that such false positives are actually quite rare, a prediction confirmed by our physiological recordings from CSTMD1. Nevertheless, inclusion of a feature designed to match the preferred target size produces robust responses, despite the motion of rich background texture in nearby parts of the receptive field.
Our model is a local target discriminator that takes no account of the presence of additional targets outside its local receptive field. By comparison, CSTMD1 is a higher-order neuron that integrates complex inputs from a large array of presumed ESTMD-like inputs. Strong evidence for long-range inhibitory interactions was revealed recently using simple paired targets (Bolzon et al., 2009). Rivalry between multiple features within the receptive field may explain some of the variability we observed.
CSTMD1 also has binocular inputs, with potent inhibition from the contralateral eye, evident from the reduction in spike rate as the target traverses the contralateral field (Fig. 1d). Bolzon et al. (2009) found that the presence of a second feature within this hemifield will suppress the response to targets presented to the excitatory receptive field. This may explain the reduction in response to the artifactual tree trunk feature (Fig. 2i, R2) in the image with the optimal target embedded, since our target would have been within the inhibitory field at the time this feature was in the receptive field center.
A further and hitherto unsuspected observation is the reduction in response to the artifactual tree trunk feature (Fig. 2d; i, R2) upon removal of the false-positive leaf feature. Whereas it is easy to see how addition of a feature (i.e., the target in Fig. 2b) could recruit the long range inhibition observed by Bolzon et al. (2009), this finding is more difficult to explain. Nordström et al. (2011) recently described additional facilitation mechanisms within the CSTMD1 receptive field, so it is feasible that the presence of the target-like group of leaves facilitated the breakthrough response to the tree trunk in Figure 2c. Although our work here provides a first step toward understanding the response of these fascinating neurons in a more natural image context, it also highlights the need for computational models that better account for the inhibitory and facilitatory interactions underlying their complex receptive fields.
Footnotes
This research was supported by the US Air Force Office of Scientific Research (FA2386-10-1-4114). We thank the manager of the Botanic Gardens of Adelaide for allowing insect collection.
References
- Barnett PD, Nordström K, O'Carroll DC. Retinotopic organization of small-field-target-detecting neurons in the insect visual system. Curr Biol. 2007;17:569–578. doi: 10.1016/j.cub.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Bolzon DM, Nordström K, O'Carroll DC. Local and large-range inhibition in feature detection. J Neurosci. 2009;29:14143–14150. doi: 10.1523/JNEUROSCI.2857-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett TS, King AJ. Vision during flight. In: Horridge GA, editor. Compound eye and vision of insects. Clarendon: Oxford UP; 1975. pp. 437–466. [Google Scholar]
- Collett TS, Land MF. How hoverflies compute interception courses. J Comp Physiol. 1978;125:191–204. [Google Scholar]
- Field DJ. Relations between the statistics of natural images and the response properties of cortical cells. J Opt Soc Am A. 1987;4:2379–2394. doi: 10.1364/josaa.4.002379. [DOI] [PubMed] [Google Scholar]
- Geurten BR, Nordström K, Sprayberry JD, Bolzon DM, O'Carroll DC. Neural mechanisms underlying target detection in a dragonfly centrifugal neuron. J Exp Biol. 2007;210:3277–3284. doi: 10.1242/jeb.008425. [DOI] [PubMed] [Google Scholar]
- Horridge GA. The separation of visual axes in apposition compound eyes. Philos Trans R Soc Lond B Biol Sci. 1978;285:1–59. doi: 10.1098/rstb.1978.0093. [DOI] [PubMed] [Google Scholar]
- Nordström K, O'Carroll DC. Feature detection and the hypercomplex property in insects. Trends Neurosci. 2009;32:383–391. doi: 10.1016/j.tins.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Nordström K, Barnett PD, O'Carroll DC. Insect detection of small targets moving in visual clutter. PLoS Biol. 2006;4:e54. doi: 10.1371/journal.pbio.0040054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K, Bolzon DM, O'Carroll DC. Spatial facilitation by a high-performance dragonfly target-detecting neuron. Biol Lett. 2011 doi: 10.1098/rsbl.2010.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D. Feature-detecting neurons in dragonflies. Nature. 1993;362:541–543. [Google Scholar]
- Olberg RM. Object- and self-movement detectors in the ventral nerve cord of the dragonfly. J Comp Physiol. 1981;141:327–334. [Google Scholar]
- Olberg RM, Worthington AH, Venator KR. Prey pursuit and interception in dragonflies. J Comp Physiol A. 2000;186:155–162. doi: 10.1007/s003590050015. [DOI] [PubMed] [Google Scholar]
- Osorio D. Mechanisms of early visual processing in the medulla of the locust optic lobe: how self-inhibition, spatial-pooling, and signal rectification contribute to the properties of transient cells. Vis Neurosci. 1991;7:345–355. doi: 10.1017/s0952523800004831. [DOI] [PubMed] [Google Scholar]
- Wiederman SD, Shoemaker PA, O'Carroll DC. A model for the detection of moving targets in visual clutter inspired by insect physiology. PLoS One. 2008;3:e2784. doi: 10.1371/journal.pone.0002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederman SD, Brinkworth RS, O'Carroll DC. Performance of a bio-inspired model for the robust detection of moving targets in high dynamic range natural scenes. J Comput Theor Nanosci. 2010;7:911–920. [Google Scholar]