FIG 2.
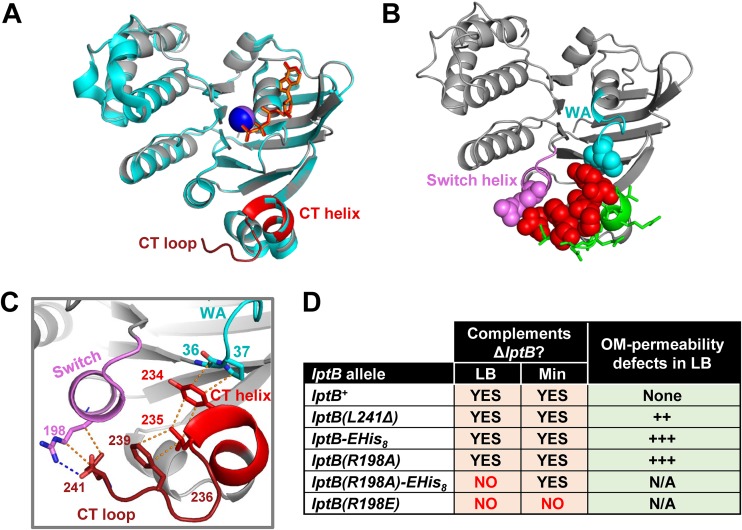
The CTD of LptB makes critical contact with the Walker A and switch helix. (A) Alignment (generated with align function in PyMol; root mean square [RMS] = 0.183) of ATP-bound structures of His8-LptBE163Q (PDB 6MBN; gray) and LptB-EHis8E163Q (PDB 4P33; cyan). The CTD of His8-LptBE163Q is colored red. In PDB 6MBN, the Na+ atom is shown as a blue sphere and ATP as orange sticks. In 4P33, the Na+ atom is shown as a purple sphere and ATP as red sticks. (B) Cartoon representation of ATP-bound LptB structure (PDB 6MBN). CTD residues that, when substituted as in the Fig. 1 legend, resulted in no defects are shown as green sticks; those that caused total or partial loss of function are shown as red spheres. Functionally important CTD residues place the CTD helix and CTD loop such that they can interact with switch helix (violet) residue R198 (violet spheres) and Walker A (cyan) residues G36 and P37 (cyan spheres). (C) Detailed interactions of the functionally important CTD (red) residues shown in panel B. Dotted lines represent polar (blue) and nonpolar (orange) interactions. Y234 interacts with G36 and P37 in the Walker A motif (cyan); Y234, L235, and F239 stack together through hydrophobic interactions; G236 is likely critical for creating a turn that both facilitates stacking of L235 and F239 and properly orients L241; L241 interacts with R198 in the switch helix that follows the switch domain. (D) Functional characterization of mutant alleles encoding changes that disrupt the interaction between position R198 of the switch helix and the CTD loop. Table data indicate the ability of lptB alleles to complement a chromosomal ΔlptB allele in rich (LB) and minimal (Min) media. Haploid strains carrying alleles that complemented in LB were tested for increased OM permeability to antibiotics by disc diffusion assay. The relative increase in OM permeability is indicated with plus signs (+). Refer to Data set S1 for data and details.