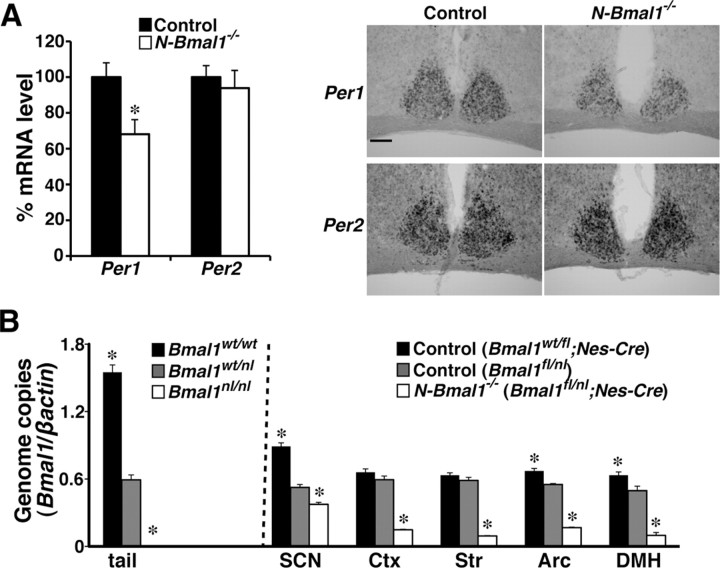
Figure 2.
Bmal1 function is mostly retained in SCN of N-Bmal1−/− mice. A, Per1 and Per2 mRNA expression levels in SCN of control mice (black bar) and N-Bmal1−/− mice (white bar) at CT8 determined by in situ hybridization. *p < 0.05 by unpaired Student's t test. Values are mean ± SEM (n = 5). Representative images are shown on the right. Scale bar, 200 μm. B, Efficiency of Cre-mediated recombination of Bmal1fl allele. Amounts of Bmal1wt and Bmal1fl alleles were quantified by real-time PCR and normalized to those of β-actin gene. A pilot analysis of genomic DNA extracted from tails (shown in leftmost field) of Bmal1w/w (black bar), Bmal1wt/nl (gray bar), and Bmal1nl/nl (white bar) mice confirmed the specificity and quantitative performance of amplification. Genomic DNA extracted from SCN, cerebral cortex, striatum, arcuate hypothalamic nucleus (Arc), and DMH of Bmal1wt/fl;Nes-Cre (black bars) and Bmal1fl/nl (gray bars) control mice and N-Bmal1−/− mice (white bars) was subjected to similar quantitative PCR. *p < 0.05 by unpaired Student's t test vs Bmal1wt/nl mice (tail) or Bmal1fl/nl mice (brain tissues). Values are mean ± SEM (n = 3 for tail analysis, n = 4 for Bmal1wt/fl;Nes-Cre mice, n = 3 for Bmal1fl/nl mice, n = 6 for N-Bmal1−/− mice). Ctx, Cerebral cortex; Str, striatum.