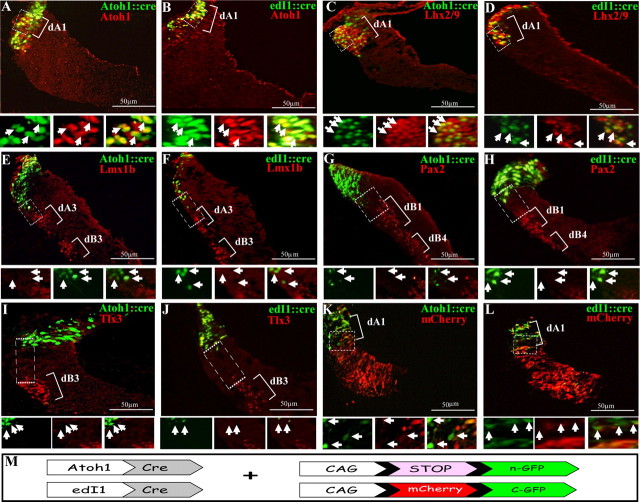
Figure 2.
Labeling of dA1 neuronal subgroup using Atoh1/edI1 enhancer elements. Cross-section views of hindbrains at the level of r4–r5 that were electroporated with Atoh1::Cre (A, C, E, G, I, K) or edI1::Cre (B, D, F, H, J, L) plasmids, along with a conditional nuclear GFP reporter (A–J) or conditional alternating mCherry/GFP (K, L) plasmids. Sections in A–J were stained with Atoh1, Lhx2/9, Lmx1b, Pax2, or Tlx3 antibodies. In all images, dA/dB subgroups are marked. Higher-magnification views of the boxed areas are represented below each panel in different channels, and arrows indicate the same cells in all channels. Plasmids and antibodies are indicated in their respective colors and scale bars are indicated. M, A scheme of the Cre/loxP-based expression systems used in the experiments. A nuclear GFP (n-GFP) is flanked by two loxP sites (pCAGG-LoxP-STOP-LoxP-nGFP), or a floxed mCherry gene is inserted between the CAGG enhancer/promoter module and the GFP gene (CAGG-loxP-mCherry-loxP-GFP). The expression of GFP in dA1 neurons is driven by Atoh1::Cre or edI1::Cre.