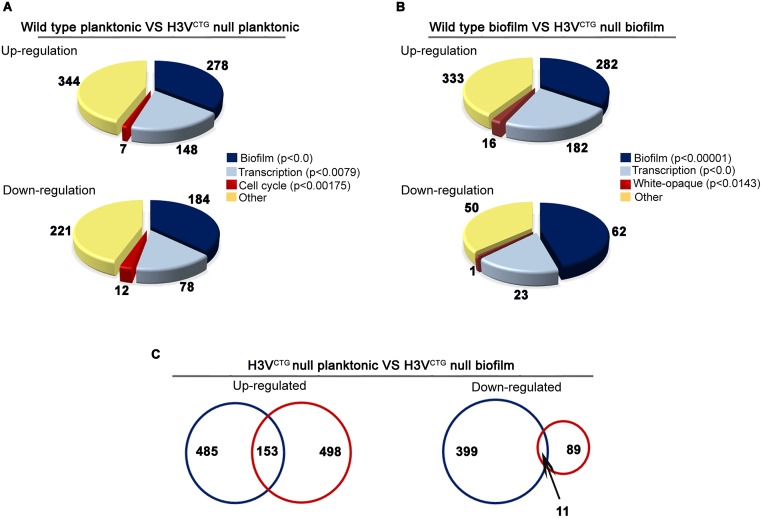
Fig 2. Loss of variant histone H3 activates the gene circuitry for biofilm development in planktonic cells.
(A) Global gene expression array analysis was performed in the wild-type and null mutants for H3VCTG grown in YPDU (planktonic mode). Functional classification of genome-wide expression data indicates that the most significantly altered pathways in the H3VCTG null mutants are biofilm, transcription, and cell cycle. (B) A similar pattern of gene expression was observed when the indicated strains were grown in Spider medium under conditions that favor biofilm growth. (C) Genome-wide expression data were compared for common genes that are altered both in planktonic and biofilm growth conditions (blue and red circles, respectively) in the H3VCTG null mutants and represented as Venn diagrams. A total of 153 up-regulated and 11 down-regulated genes are common between these 2 data sets. YPDU, yeast peptone dextrose supplemented with uridine.