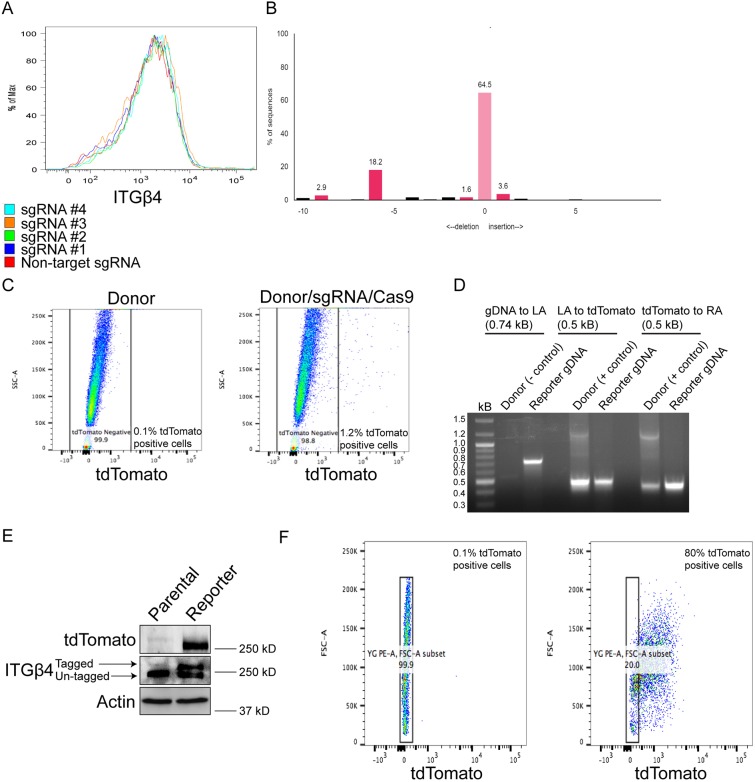
Fig. 1.
Design, approach and validation for Crispr/Cas9-mediated integrin β4 tagging. (A) Comma-d1 cells expressing Cas9 and either a non-target sgRNA or one of four sgRNAs targeting the region of the last exon of mouse integrin β4 were processed for flow cytometry to determine the effects on integrin β4 surface abundance. (B) Genomic DNA was isolated from cells sorted as in A and was processed for TIDE analysis to quantify the cutting efficiency of each sgRNA. The y-axis indicates the percentage of sequences altered. The predicted Cas9 cut-site is indicated by ‘0’ in the x-axis, which also depicts other sites away from the predicted Cas9 cut-site that have insertions or deletions. (C) Comma-d1 cells were transfected with donor plasmid alone, or donor plasmid, sgRNA #2 and Cas9 plasmid, and processed for flow cytometry to quantify the proportion of tdTomato-positive cells, which were subsequently processed for single-cell sorting. (D) Genomic DNA from clones as described in C was isolated and processed for PCR to determine the correct genomic insert of tdTomato. (E) Immunoblotting was performed to confirm expression of tdTomato and the heterozygous tdTomato-tagged integrin β4 allele. (F) Expression of tdTomato was quantified by flow cytometry to determine the percentage of tdTomato-positive cells.