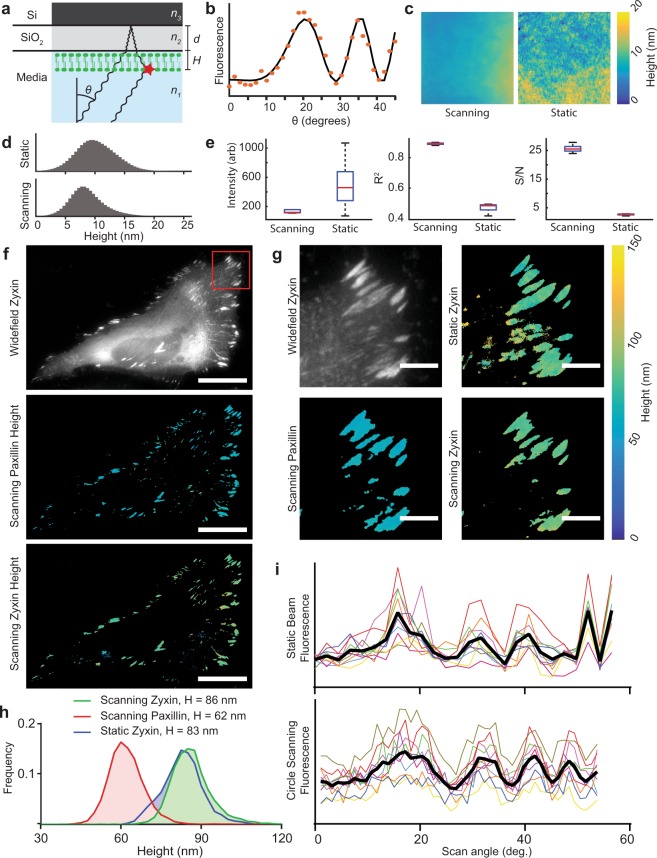
Figure 3.
Azimuthal beam scanning reduces incidence angle dependent laser artifacts in scanning angle interference microscopy (SAIM) imaging. (a) Schematic illustration of SAIM. Samples were prepared on reflective silicon substrates with a defined oxide layer of thickness d and excited with a laser at incidence angle θ to generate an axial interference pattern with known spatial frequency. The measured fluorescence intensity at distance from the substrate H varies with θ for a supported lipid bilayer (SLB) labeled with DiI. (b) Example single-pixel raw data (orange dots) and best-fit curve. (c) Reconstructed SLB height maps with circle scanning or static beam excitation schemes (full image 147.9 × 147.9 μm). (d) Pooled DiI height distributions in SLBs acquired with static beam and circle scanning excitation schemes (n = 5 regions, 4.2 × 106 pixels per region). (e) Quantification of the average fit parameters in circle scanning and static beam SAIM imaging of the 5 lipid bilayer regions in d. Red lines indicate mean, boxes the central quartiles, whiskers min and max. (f) Widefield and circle scanning SAIM height reconstructions of a live HeLa cell expressing the focal adhesion components mEmerald-Zyxin and mCherry-Paxillin, scale bars 25 μm. (g) Enlarged view of the region boxed in f. The colored images represent pixel-by-pixel SAIM height reconstructions of mEmerald-Zyxin or mCherry-Paxillin as noted, scale bars 5 μm. A static beam reconstruction of zyxin height is included for comparison. (h) Distribution of pixelwise height fits from adhesion complexes in g. Heights are the average of all non-zero fits. (i) Plots of fluorescence intensity as a function of scan angle for the static and circle scanning Zyxin images in g. Colored lines indicate the same 10 representative pixels from each image, black lines the average of all 10 pixels.