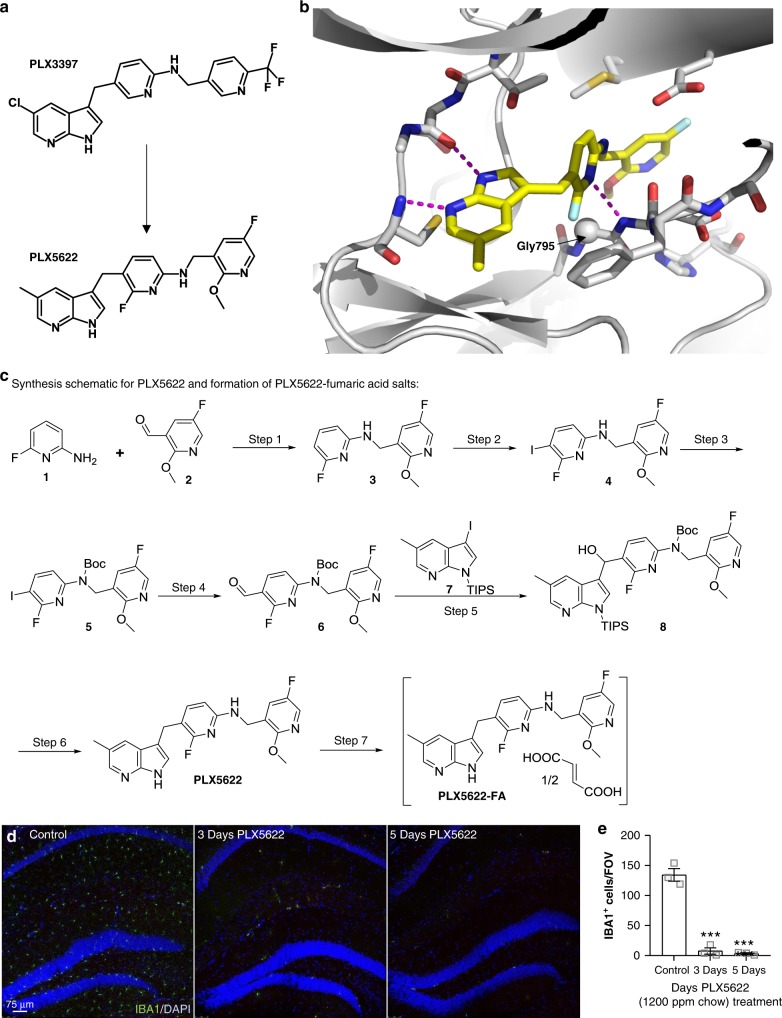
Fig. 2.
Design and synthesis schematic of the CSF1R inhibitor PLX5622 for extended microglial elimination. a Chemical structure of selective CSF1R inhibitor PLX5622. PLX5622 is structurally similar to CSF1R/KIT/FLT3 inhibitor PLX3397 with modifications concentrated on the two pyridine moieties. b X-ray crystal structure of the CSF1R-PLX5622 complex. PLX5622 is anchored to the active site of CSF1R by 3 hydrogen bonds (indicated by dashed lines). 7-Azaindole of PLX5622 forms two hydrogen bonds with the hinge region of CSF1R whereas the central pyridine ring forms a hydrogen bond with the main chain amino group of Phe796. The CSF1R selectivity is largely determined by the interaction between PLX5622 and Gly795 (represented as a sphere), which is a bulkier residue (cysteine) in KIT and FLT3. The substitutions on the tail pyridine ring also contribute to the selectivity. This group displaces the juxatemembrane (JM) region (absent in the structure) from the allosteric site. In contrast, PLX3397 binds CSF1R in the presence of the JM region. c Synthesis schematic for PLX5622 and subsequent formation of PLX5622-fumaric acid salt. PLX5622 was synthesized from commercially available 2-amino-6-fluoropyridine (1), 5-fluoro-2-methoxypyridine-3-carbaldehyde (2), and 3-iodo-5-methyl-1-(triisopropylsilyl)-1H-pyrrolo[2,3-b]pyridine (7) using the reaction scheme. d Immunolabeling of microglia (IBA1 in green) and cell nuclei (DAPI in blue) of two-month-old animals treated with control or 1200 ppm PLX5622-formulated chow. Scale bar = 75 μm. E, Quantification of hippocampal microglial number over 5 days of treatment with PLX5622 (p < 0.001 for 3d and 5d). Two-way ANOVA with Dunnet’s post hoc test. n = 3 for Control, n = 3 for 3d PLX5622, n = 3 for 5d PLX5622. Statistical significance is denoted by ***p < 0.001. Error bars indicate SEM