Fig. 9.
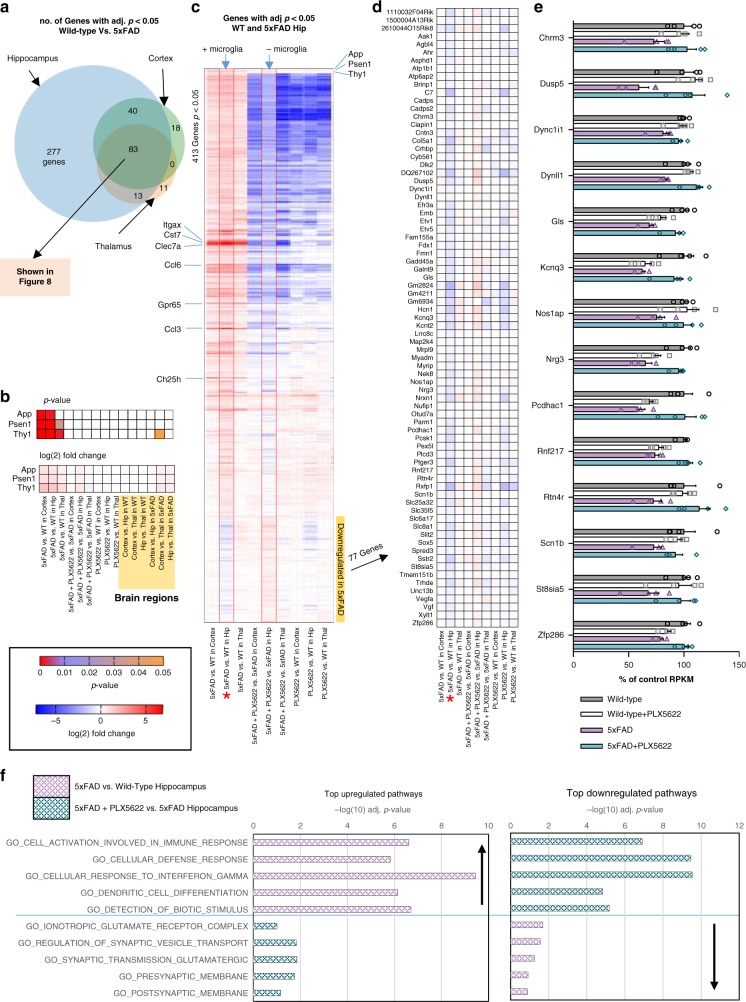
Microglia mediate downregulation of neuronal/plasticity genes in the hippocampus in response to AD pathology. a Venn diagram showing the number of differentially expressed genes (adjusted (adj.) p < 0.05) for cortex, hippocampus, and thalamus for wild-type (WT) vs. 5xFAD mice. b Heatmap of the adj. p-value and log(2) fold change for all 9 comparison groups (5xFAD vs. WT in cortex, hippocampus, or thalamus, 5xFAD + PLX5622 vs. 5xFAD in cortex, hippocampus, or thalamus, and WT + PLX5622 vs. WT in cortex, hippocampus, or thalamus), as well as 3 comparisons between brain regions for both WT and 5xFAD mice, for the transgene components in 5xFAD mice (App, Psen1, Thy1). c Heatmap of all 413 gene expression differences identified in the hippocampus for wild-type vs. 5xFAD, expressed as log (2) fold change, and all 9 comparison groups included. Hippocampus for 5xFAD vs. WT and 5xFAD + PLX5622 vs. 5xFAD highlighted by red border, showing that most significant gene expression changes induced by pathology do not occur in the absence of microglia. d Downregulated genes in the hippocampus from (a), displayed as a heatmap of log(2) fold change differences for all 9 comparisons, showing that the same genes are not downregulated in the absence of microglia (5xFAD + PLX5622 vs. 5xFAD Hip). e RPKM values plotted on a log(2) scale for a subset of plasticity genes from (d). f Five significantly upregulated and downregulated pathways for 5xFAD vs. wild-type hippocampus (red), along with respective -log(10) p-values plotted: most upregulated pathways are related to immune function, while downregulated pathways are mainly associated with neuronal and synaptic activity. The same pathways are displayed for the comparison between 5xFAD + PLX5622 vs. 5xFAD hippocampus (yellow), showing that the absence of microglia prevents the upregulation of immune pathways and the downregulation of synaptic pathways. Expression difference denoted by *. Log(2) fold change and p-values indicated by respective scale bar. n = 4 per group for all analyses. Error bars indicate SEM