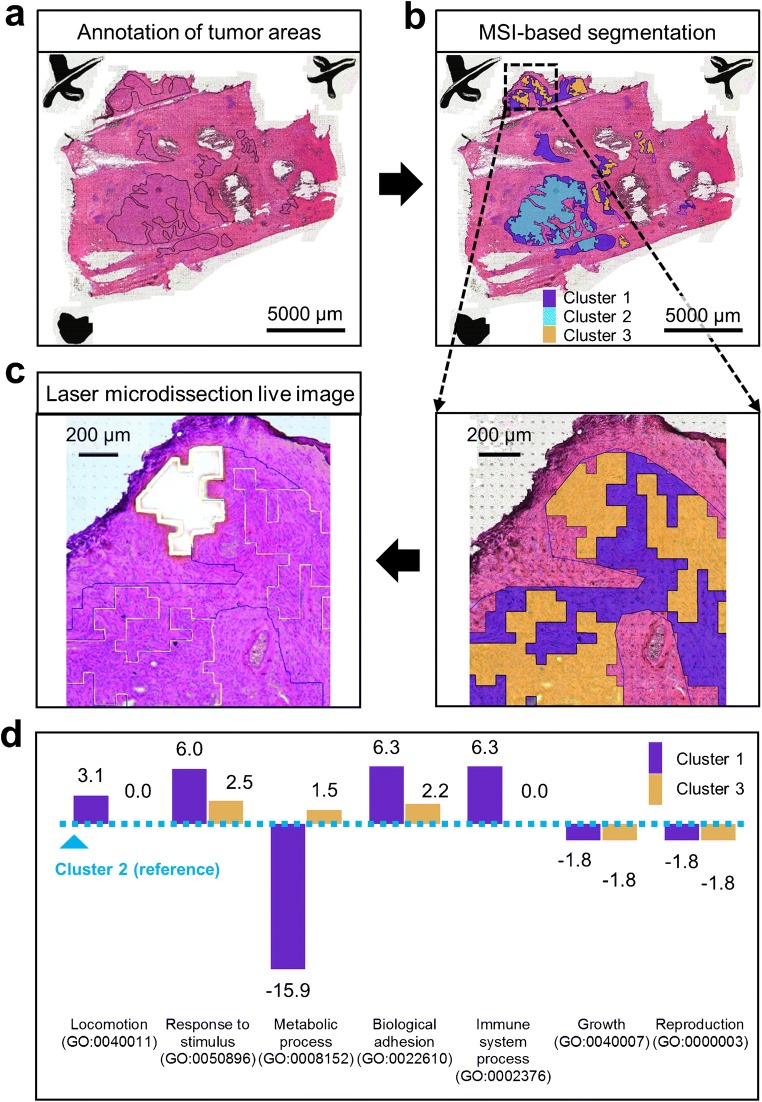
Fig. 2.
Laser microdissection of MSI-defined intratumor segments subsequently characterized by microproteomics. (a) First, tumor areas were annotated by a pathologist on the H&E image. (b) Then, mass spectrometry imaging (MSI) lipid data of the tumor was used to spatially segment the tumor areas into three clusters using non-negative matrix factorization. After image processing of the segments including smoothing and boundary detection (ESM, Figs. S3 and S4), the segmentation image was upscaled to the resolution of the H&E image. (c) The segments’ boundaries were finally transferred to the LMD software using the previously established co-registration pipeline. Each MSI cluster was then microdissected by the LMD system for the subsequent microproteomics analysis. This resulted in the identification and label-free quantification of over 1000 common proteins. Cluster exclusive over- and under-expressed proteins were submitted to gene ontology analysis (ESM, Fig. S5). (d) shows selected differences in molecular functions between the clusters (in percentage points with respect to cluster 2)