Figure 1.
Mono-association with LpWJL Buffers Phenotypic and Transcriptomic Variation during Growth and Development in the DGRP Lines
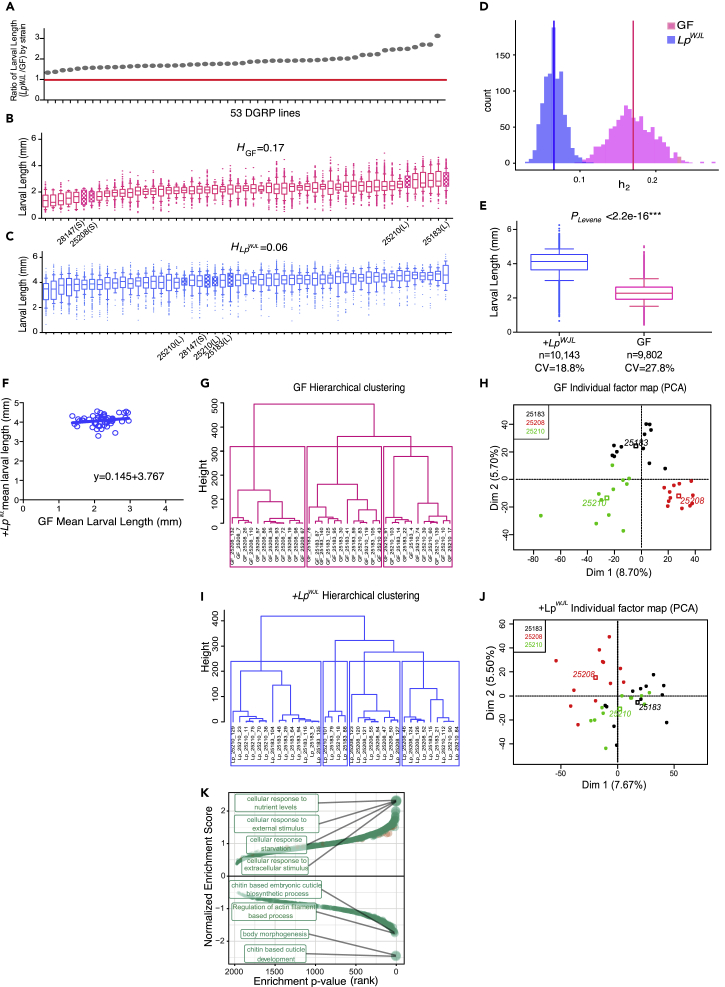
(A) The ranking of larval growth gain of 53 DGRP lines was used for GWAS to uncover host variants associated with growth benefits conferred by LpWJL. Each gray dot represents the quotient of average mono-associated larval length (Figure 1C) divided by the average length of GF larval length (Figure 1B) from each DGRP line on Day 7 AEL (after egg lay). The red line marks the ratio of “1,” indicating that all tested DGRP lines benefited from LpWJL presence.
(B and C) The average larval length on Day 7 AEL for each of the 53 DGRP lines. (Data are represented as mean and 10–90 percentile. Unless specified, all box plots in this manuscript present the same parameters.) Each line in the box represents the average length from pooled biological replicates containing all viable larvae from all experimental repeats. From each strain, there are between 10 and 40 viable larvae in each replicate, 3 biological replicates for each experiment, and 2 to 3 repeats of the experiments. (B): germ-free (GF, pink), (C): mono-associated (+LpWJL, blue). Note the heritability estimate (H) in the GF population is higher than in the mono-associated population (17% versus. 6%). The filled boxes denote the “small (S)” and “large (L)” DGRP lines that were selected for setting up the F2 crosses (see Figure S3A for crossing schemes).
(D) The estimation of empirical distribution of heritability indices in GF and LpWJL mono-associated larvae (p < 2.2 × 10−16, Kolmogorov-Simirnov test). The vertical lines are reported H values.
(E) Box and whiskers plots showing average larval length derived from pooled GF (pink) or LpWJL (blue) mono-associated DGRP lines. The coefficient of variation in the GF population (27.82%) is greater than that of the mono-association population (18.74%). Error bars indicate 10th to 90th percentile. Levene's test is used to evaluate homocedasticity and Mann-Whitney test for difference in the median (***p < 0.0001).
(F) Scatterplot to illustrate that LpWJL buffers size variation in ex-GF larvae in the DGRP population. Each data point represents the intercept of the average GF larvae length and its corresponding mono-associated average length at Day 7 for each DGRP line. If genetic variation was the only factor influencing growth in both GF and mono-associated flies, the slope of the scatterplot should theoretically be 1 (Null hypothesis: slope = 1. p < 0.0001: the null hypothesis is therefore rejected. A linear standard curve with an unconstrained slope was used to fit the data).
(G–J) Hierarchical clustering (G: GF and I: mono-associated) and principal-component analyses (PCA) (H: GF and J: mono-associated) based on individual larvae transcriptome analyses show that the samples cluster more based on genotypes when germ-free (G and H, G: Pgenotype = 1.048 × 10−8, H: R2 Dim1 = 0.73, Pgenotype = 7.81 × 10−10, R2Dim2 = 0.72, Pgenotype = 1.12 × 10−9) than mono-associated (I and J, I: Pgenotype = 0.000263, J: R2Dim1 = 0.42, Pgenotype = 0.00017, R2Dim2 = 0.31, Pgenotype = 0.00269). A scaled PCA using the genotype as categorical supplementary variable was performed. A hierarchical clustering on principle components (HCPC) was applied on the PCA results, and the trees were automatically cut based on inertia drop (Figure S2F). Both PCA and HCPC were performed with the R package FactoMineR on the voom corrected read counts. Correlations between the genotype variable and PCA dimensions or HCPC clusters were assessed by χ2 tests. The dots represent the different samples according to genotype, and the empty squares are the calculated centers for each genotype.
(K) Gene set enrichment analysis based on the change in standard deviation of gene expression. Positive enrichment indicates gene sets that are enriched in the genes whose expression level variation increases in response to LpWJL mono-association. Negative gene sets are those that are enriched in the genes whose expression level variation decreases in response to LpWJL mono-association. The top four positively and negatively enriched sets are labeled. The genes whose expression levels are reduced by LpWJL mono-association predominantly act in chitin biosynthesis and morphogenesis (see also Figure S2).