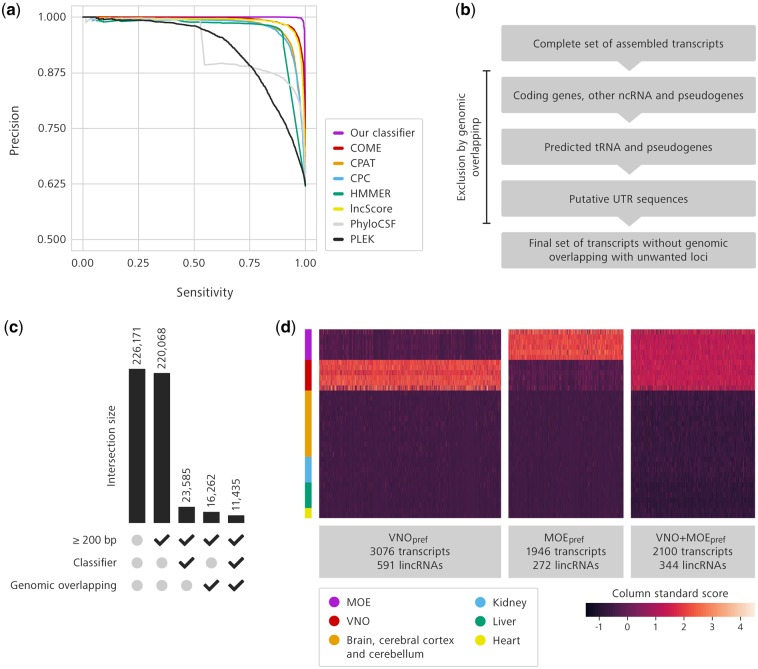
Figure 1.
An improved pipeline identifies hundreds of long non-coding RNAs preferentially expressed in the olfactory organs. (a) Precision-recall curve of our lncRNA classifier (purple) in comparison with other currently available classification models. (b) Schematic representation of the sequential removal of transcripts displaying genomic overlap with unwanted genomic loci. (c) Bar plot representing the filtering steps for lncRNA identification. Each bar represents the number of transcripts kept by the combination of sieves represented by the check marks below (selection of long transcripts, selection of transcripts that pass our classifier, and selection of transcripts that do not overlap with unwanted genomic loci). The numbers of filtered transcripts are shown above each bar. (d) Heatmap of expression values for lncRNAs preferentially expressed in the MOE (MOEpref), in the VNO (VNOpref), or in both olfactory organs (VNO+MOEpref). Abundance values were normalized by standardized score calculation, which represents deviation of expression from the average. Columns are transcripts and lines are distinct RNA-Seq libraries from tissues represented by the colour code on the left.