Abstract
Sirtuin 1 (SIRT1) is a histone deacetylase implicated in stem cell homeostasis. Conditional Sirt1 deletion in the hematopoietic stem and progenitor system promotes hematopoietic stem and progenitor cell (HSPC) expansion under stress conditions. In addition, SIRT1 activators modulate the capacity and HSPC numbers in the bone marrow (BM). To investigate the role of SIRT1 in the BM niche, a conditional Sirt1 deletion in the BM niche was generated in a mouse model for the present study. Multicolor flow cytometric analyses were performed to determine HSC cell populations. Using 5-fluorouracil-induced proliferative stress, a survival curve was produced. In the present study, Sirt1 deletion in the BM niche demonstrated that the production of mature blood cells, lineage distribution within hematopoietic organs and frequencies of HSPC populations were comparable to those of controls. Additionally, Sirt1 deletion in the BM niche did not perturb HSC maturation under stress induced by transplantation. Therefore, these observations suggest that SIRT1 serves a dispensable role in HSC maturation in the BM niche.
Keywords: sirtuin 1, bone marrow niche, hematopoietic stem cells
Introduction
The bone marrow (BM) is a complex tissue comprised of multiple subsets of stromal cells. Studies using a number of different experimental approaches are beginning to unravel the stromal cell profile in the BM (1). The BM niche was the first proposed and experimentally defined niche for stem cells in mammals (2). Bone marrow stromal cells serve to regulate hematopoiesis in the hematopoietic BM niche (1–3). Hematopoietic stem cells (HSC) mainly reside in the BM niche and participate in tissue regeneration under hematopoietic stress such as transplantation (4). However, under stress condition including transplantation, these HSCs can lose their capacity for self-renewal, resulting in stem cell exhaustion. Under these circumstances, allogenic HSCs have to be engrafted into the BM niche and subsequently expanded in a process known as transplantation (5).
The histone deacetylase sirtuin 1 (SIRT1) regulates a number of cellular responses, including cell proliferation, apoptosis and inflammatory responses by protein deacetylation (6–8). In particular, SIRT1 has also been implicated in stem cell homeostasis (9–12). Previous studies suggest that SIRT1 serves a role in HSC maturation and lineage specification as demonstrated by a conditional deletion approach, where Sirt1 expression was knocked out only in the hematopoietic system (13,14). However, little is known about the role of SIRT1 in the BM niche, since SIRT1 was found to be dispensable for HSC activity due to developmental HSC adaptation in surviving conventional Sirt1 KO mice in another study (15). In addition, the SIRT1 activator Resveratrol modulates HSC capacity in the BM in vivo (16), whereas SRT3025, another SIRT1 activator, also altered the number of HSCs (17); these observations suggest that SIRT1 is important for HSC maturation. In the present study aim was to elucidate the role of SIRT1 in the BM niche to provide novel insight for HSC transplantation.
Materials and methods
Ethics statement and animals
All animals were housed in an air-conditioned room (22–25°C; 12-h light/dark cycle; 50% humidity) with free access to food pellets and tap water. Ocn-Cre transgenic mice used in BM niches (18) and Sirt1 mice (19) have been described (Fig. S1). Ocn-Cre, floxed Sirt1 and wild-type C57BL/6 mice were purchased from Jackson Laboratory and employed for experiments. The Ocn-Cre mice are widely used to target osteoblasts in the BM niche (18). To investigate the role for HSC maturation of Sirt1 in the BM niche, such as in osteoblast, Sirt1floxed/floxed mice were crossed with Ocn-Cre. A total of 70 mice (female; weight, 15–18 g; age, 8–10 weeks) were used for experiments.
Transplantations of BM cells were performed as described previously (20). Briefly, mice were euthanized then tibias and femurs were recovered. Scalpel was used to cut the ends of bone off and then a syringe with 27G needle (filled with PBS supplemented with 2% FBS and 1% P/S) was used to flush into a 50 ml tube covered with 40 µM nylon filter. Recovered BM cells were lysed in RBC lysis buffer (Lonza Group, Ltd.) for 5–10 min on ice. Samples were then washed with 10 ml PBS supplemented with 2% FBS and 1% P/S and centrifuged 5 min at 1,500 rpm. Supernatant was aspirated and the cell pellets were collected for the experiments.
Control and Sirt1Δ/Δ mice received lethal doses of irradiation (9.5 Gy, Gammacell 3000) prior to the transplantation of 200 µl 1×106 BM cells by retro-orbital injection on the same day as previously described (21). For serial 5-FU treatments, 5-FU (150 mg/kg) was injected intraperitoneally every seven days until 100% animal mortality was achieved (22). For the duration of the present study, the following criteria were applied to define a mouse as moribund, which would require immediate euthanasia (23,24): i) Ruffled and/or matted fur; ii) weight loss of >15%; iii) hypothermia (detected by touching); iv) hunched posture; v) unable to eat or drink freely; vi) barrel rolling. No animals satisfied these criteria in the present study.
Reverse transcription-quantitative PCR (RT-qPCR)
Reverse transcription and quantitative PCR were performed as previously described (14,25). Briefly, total RNAs were extracted from BM cells using QIAGEN RNeasy-Plus Mini-columns according to manufacturer's protocol (Qiagen, Inc.). cDNA synthesis were performed following the protocols of the manufacturer of the kit (Bio-Rad Laboratories, Inc.). qPCR was performed with SYBR Green Mix (Roche Diagnostics) and LightCycler 96 instrument (Roche Diagnostics) and data were normalized to the housekeeping gene GAPDH. The mRNA levels were measured using the 2−ΔΔCq method of quantification (26). RT-qPCR was performed using the following primers: Sirt1 forward, 5′-CTGAAAGTGAGACCAGTAGCA-3′ and reverse, 5′-GATGAGGCAAAGGTTCCCTA-3′ and GAPDH forward, 5′-GCACAGTCAAGGCCGAGAAT-3′ and reverse, 5′-GCCTTCTCCATGGTGGTGAA-3′.
Flow cytometric analysis
Flow cytometry was performed as described previously (20). Briefly, BM cells were collected from femurs and tibias of the mice by flushing using fluorescence-activated cell sorting buffer, which consisted of phosphate buffered saline containing 2% fetal bovine serum (Hyclone; GE Healthcare Life Sciences) and 0.1% sodium azide. Peripheral blood cells were collected from the tail vein. Flow cytometry was performed using antibodies listed in Table S1. Data acquisition and analysis were performed with Cell Quest v.3.3 or Diva software v.6.1.3 (BD Biosciences) and with FlowJo software v.10.3 (FlowJo LLC) respectively.
Statistical analysis
The statistical significance of differences in the means between two populations were assessed using two-tailed unpaired Student's t-test. P<0.05 was considered to indicate a statistically significant difference for pairwise comparisons (between indicated genotypes in the same population). The Kaplan-Meier log-rank test was used to analyze survival data.
Results
Effects of Sirt1 deletion on the BM niche
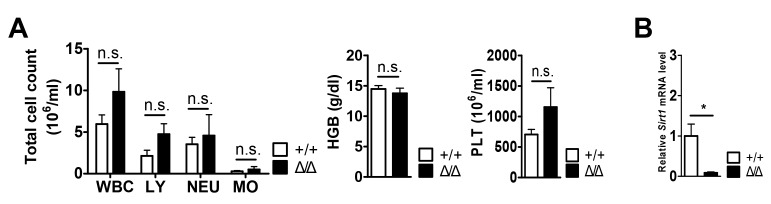
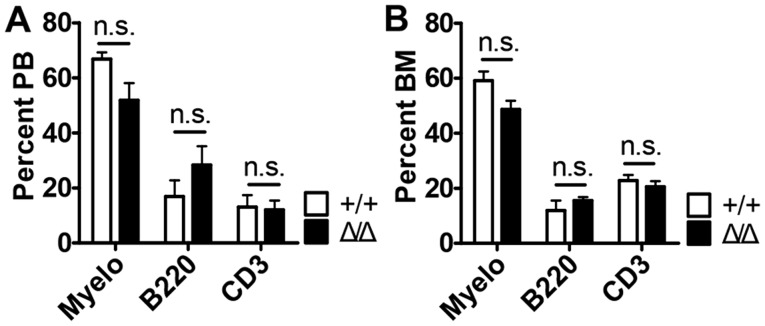
SIRT1 is a critical regulator of homeostatic adult HSCs in mice (13,14). Based on these observations, it is tempting to speculate that SIRT1 may also serve a role for HSC maturation in the BM niche. To investigate the role for HSC maturation of SIRT1 in the BM niche, Sirt1floxed/floxed mice were crossed with Ocn-Cre transgenic mice, resulting in Sirt1Δ/Δ mice. A previous study showed that the loss of Sirt1 in the hematopoietic system resulted in anemia and specified lineage decision (myeloid versus lymphoid lineage specification) (14). However, Sirt1 deletion in the BM niche demonstrated no differences in the numbers of white blood cells (WBCs), lymphocytes (LY), neutrophils (NEU), monocytes (MO), hemoglobin (HGB), and platelets (PLT) (Fig. 1A). It was also found that Sirt1Δ/Δ mice did not result in significant differences compared with wild-type mice in their ability to produce mature myeloid cells (Mac1+ cells), T cells (CD3+ cells) and B cells (B220+ cells) in the peripheral blood (PB) and bone marrow (BM) (Fig. 2 and Fig. S2). The expression of Sirt1 was decreased in the CD31−CD45−Ter119− BM niche cells of Sirt1Δ/Δ mice compared with Sirt1+/+ mice, which confirmed Sirt1 knockdown (Fig. 1B).
Figure 1.
No changes in white blood cells, hemoglobin, and platelets counts of Sirt1Δ/Δ mice. (A) White blood cell counts, lymphocytes, neutrophil, monocytes, hemoglobin and platelet measurements from mice of the indicated genotype (n=4–5). (B) Reverse transcription-quantitative PCR analysis of Sirt1 expression in CD31−CD45−Ter119− Sirt1Δ/Δ bone marrow cells (n=3). Data are presented as mean + SEM. *P<0.05. n.s., not significant; Sirt1, sirtuin 1; Sirt1Δ/Δ, sirtuin 1 conditional knockout; WBC; white blood cells; LY, lymphocytes; NEU, neutrophils; MO, monocytes; HGB, hemoglobin; PLT, platelet; CD, cluster of differentiation.
Figure 2.
Sirt1 deletion in the BM niche is dispensable for maintaining mature hematopoietic lineage cells. (A and B) The percentages of myeloid, B and T cells from (A) the peripheral blood, and (B) the bone marrow of Sirt1Δ/Δ and control mice (n=4–5). Data are presented as mean + SEM. Mac1, macrophage-1 antigen; n.s., not significant; Sirt1, sirtuin 1; Sirt1Δ/Δ, sirtuin 1 conditional knockout; Myelo, Myeloid (Mac1+) cells; B220+ cells, B cells; CD3+ cells, T cells; PB, peripheral blood; BM, bone marrow.
Effects of Sirt1 loss on HSPCs maturation were examined
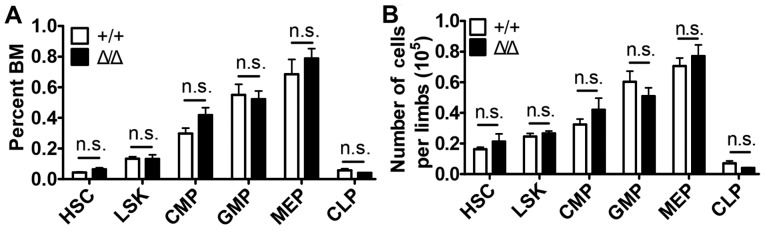
No differences in the numbers of HSCs (Lin−Sca1+cKit+CD150+CD48−, SLAM cells), Lin−Sca1+cKit+ (LSK) progenitor cells, common myeloid progenitor (CMP) cells (Lin−Sca1−cKit+CD34+CD16/32−), granulocyte-macrophage progenitor (GMP) cells (Lin−Sca1−cKit+CD34+CD16/32+), megakaryocyte-erythroid progenitor (MEP) cells (Lin−Sca1−cKit+CD34−CD16/32−) and common lymphoid progenitor (CMP) cells (Lin−Sca1lowcKitlowCD127+) were observed in the Sirt1Δ/Δ mice compared with control mice (Fig. 3 and Fig. S3).
Figure 3.
Sirt1 deletion in the BM niche is dispensable for maintaining hematopoietic stem and progenitor cells. (A) The frequencies and (B) number of cells/limb of HSC, LSK, CMP, GMP, MEP and CLP; from the BM of Sirt1Δ/Δ and control mice (n=4–5). Data are presented as mean + SEM. n.s., not significant; Sirt1, sirtuin 1; Sirt1Δ/Δ, sirtuin 1 conditional knockout; Lin, lineage; Sca1, spinocerebellar ataxia type 1; cKit, mast/stem cell growth factor receptor kit; HSC, hematopoietic (Lin−Sca1+cKit+CD150+CD48−) stem cells; LSK, Lin−Sca1+cKit+ cells; CMP, common myeloid progenitor (Lin−Sca1−cKit+CD34+CD16/32−) cells; GMP, granulocyte-macrophage progenitor (Lin−Sca1−cKit+CD34+CD16/32+) cells; MEP, megakaryocyte-erythroid progenitor (Lin−Sca1−cKit+CD34−CD16/32−) cells; CLP, common lymphoid progenitor (Lin−Sca1lowcKitlow CD127+) cells. BM, bone marrow.
Sirt1 deletion in the BM niche does not alter HSC maturation
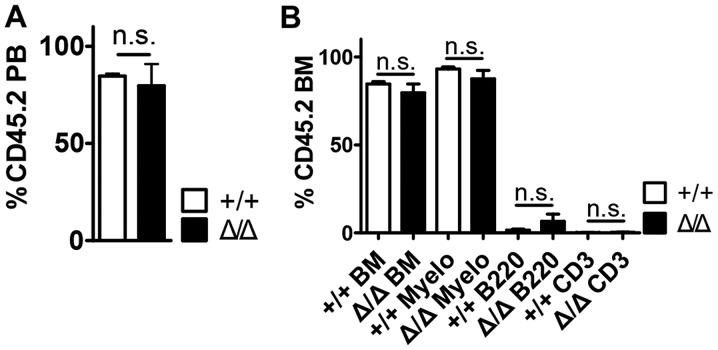
To assess the role of HSC maturation following Sirt1 deletion in the BM niche under stress condition, wild-type BM cells were transplanted into Sirt1Δ/Δ mice, which had been lethally irradiated to remove endogenous blood cells (Fig. 4 and Fig. S4). Compared with control mice, Sirt1Δ/Δ mice exhibited no detectable effects on peripheral blood (PB) donor chimerism (Fig. 4A), bone marrow (BM) donor chimerism and the numbers of mature hematopoietic cell types up to 16 weeks post-transplantation (Fig. 4B). In addition, Sirt1Δ/Δ mice demonstrated no differences in the numbers of WBCs, Lys, NEUs, MOs and the levels of hemoglobin (HGB) four weeks post-transplantation compared with control mice (Fig. S5).
Figure 4.
Hematopoietic stem cells were preserved in Sirt1Δ/Δ mice under transplantation stress. (A) Peripheral blood was collected and analyzed for CD45.2+ chimerism at 4 months post-transplantation (n=5). (B) The frequencies of myeloid, B and T cells (CD3+) in the BM from transplant recipients are shown 4 months post-transplantation (n=5). Data are presented as mean + SEM. n.s., not significant; Sirt1, sirtuin 1; Sirt1Δ/Δ, sirtuin 1 conditional knockout; Mac1, macrophage-1 antigen; Myelo, Myeloid (Mac+) cells; B220, B cells; CD3, T cells; PB, peripheral blood; BM, bone marrow.
Next the Sirt1Δ/Δ mice were repeatedly treated with 5-FU weekly to assess their sensitivity to stress under non-transplantation stress. Exposure of Sirt1Δ/Δ mice to serial proliferative stress induced by 5-FU (22) exerted in no significant differences in animal mortality compared with control mice, according to the Kaplan-Meier curve (Fig. 5).
Figure 5.
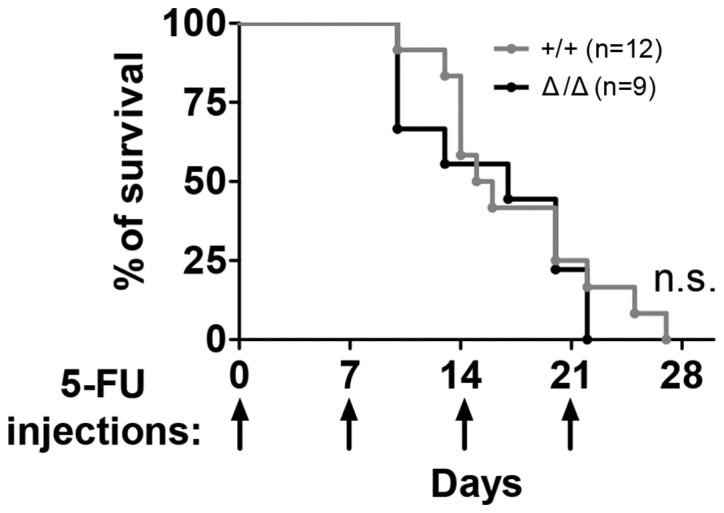
Sirt1Δ/Δ mice did not respond differently to stress induced by 5-FU compared with control. Survival of Sirt1Δ/Δ mice following weekly 5-FU (150 mg/kg) injections (n=9–12). Arrows indicate the day of 5-FU injection. n.s., not significant; Sirt1, sirtuin 1; Sirt1Δ/Δ, sirtuin 1 conditional knockout; 5-FU, 5-fluorouracil.
Discussion
In the present study, it was demonstrated that Sirt1 deletion in the BM niche (Sirt1Δ/Δ) was dispensable for maintaining mature hematopoietic lineage cells and HSC maturation. Additionally, HSCs were preserved in Sirt1Δ/Δ under transplantation stress. Also, Sirt1Δ/Δ mice did not respond differently to 5-fluorouracil (5-FU)-induced stress compared with control mice. Collectively, these findings suggested that Sirt1 deletion in the BM niche is dispensable for maturation of HSC.
First, a previous study showed that Sirt1 deletion in the HSCs resulted in anemia, a significantly decreased number of lymphocytes and increased numbers of neutrophils, monocytes and eosinophils (16). SIRT1 is essential for the myeloid versus lymphoid lineage specification in the hematopoietic blood cells (14). However, in the present study it was found that Sirt1 deletion in the BM niche lead to normal production of mature blood cells and lineage distribution within BM cells. In addition, no differences were observed between Sirt1Δ/Δ mice and wild-type mice in their ability to produce mature cells in the PB and BM.
A previous study showed that HSC expanded in response to the loss of Sirt1 in the hematopoietic compartment (13), suggesting that SIRT1 was dispensable for HSC activity due to the developmental adaptation of HSCs (15). The question on the role of SIRT1 in HSC maturation has long been debated. However, the present study found that Sirt1 deletion in the BM niche demonstrated pools of HSPC populations that were comparable to those of controls.
Finally, previous studies showed that SIRT1 is essential for HSC maturation under stress conditions, including resveratrol treatment and Fanconi anemia, SIRT1 activators enhanced HSPC capacity of the BM in vivo and increased the number of HSCs (Lin−Sca1+cKit+ cells and Lin−Sca1+cKit+CD150+CD48− cells) (16,17). However, in the present study it was found that Sirt1Δ/Δ deletion in the BM niche did not perturb HSC maturation under transplantation stress or in the presence of 5-FU, unlike Sirt1 deletion in the HSCs (13,14). Collectively, SIRT1 serves a dispensable role in HSC maturation in the BM niche, but not in the hematopoietic compartment. The findings of the present study contributed to better understanding of the molecular biology involved in the BM niche and hematopoiesis.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (grant no. NRF-2018R1C1B6001290) and the Convergence Medical Institute of Technology R&D project (grant no. CMIT2019-03), Pusan National University Hospital.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
SMP, DHSh, JYK, DYP, DHSo, YHK, SMK, JHK, SSB, KK, CDKi, CDKa and DL contributed to the design of the study. SMP, JYK and CMH acquired and analyzed the data. SMP and DL drafted the manuscript. DHSh, JYK, DYP, DHSo, YHK, SMK, JHK, SSB, KK, CDKi, CDKa and DL revised and edited the manuscript. DL acquired funding and resources. All authors read and approved the final version of the manuscript.
Ethical approval and consent to participate
The study was approved by the Ethics Committee of Pusan National University School of Medicine, Republic of Korea.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- 2.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Xie T. Stem cell niche: Structure and function. Annu Rev Cell Dev Biol. 2005;21:605–631. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 4.Trumpp A, Essers M, Wilson A. Awakening dormant haematopoietic stem cells. Nat Rev Immunol. 2010;10:201–209. doi: 10.1038/nri2726. [DOI] [PubMed] [Google Scholar]
- 5.Thomas ED. Bone marrow transplantation from the personal viewpoint. Int J Hematol. 2005;81:89–93. doi: 10.1532/IJH97.04197. [DOI] [PubMed] [Google Scholar]
- 6.Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins-novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 7.Park SY, Lee SW, Kim HY, Lee SY, Lee WS, Hong KW, Kim CD. SIRT1 inhibits differentiation of monocytes to macrophages: Amelioration of synovial inflammation in rheumatoid arthritis. J Mol Med (Berl) 2016;94:921–931. doi: 10.1007/s00109-016-1402-7. [DOI] [PubMed] [Google Scholar]
- 8.Park SY, Lee SW, Lee SY, Hong KW, Bae SS, Kim K, Kim CD. SIRT1/adenosine monophosphate-activated protein kinase α signaling enhances macrophage polarization to an anti-inflammatory phenotype in rheumatoid arthritis. Front Immunol. 2017;8:1135. doi: 10.3389/fimmu.2017.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou X, Chae HD, Wang RH, Shelley WC, Cooper S, Taylor T, Kim YJ, Deng CX, Yoder MC, Broxmeyer HE. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood. 2011;117:440–450. doi: 10.1182/blood-2010-03-273011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh SK, Williams CA, Klarmann K, Burkett SS, Keller JR, Oberdoerffer P. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J Exp Med. 2013;210:987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rimmele P, Bigarella CL, Liang R, Izac B, Dieguez-Gonzalez R, Barbet G, Donovan M, Brugnara C, Blander JM, Sinclair DA, Ghaffari S. Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Reports. 2014;3:44–59. doi: 10.1016/j.stemcr.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leko V, Varnum-Finney B, Li H, Gu Y, Flowers D, Nourigat C, Bernstein ID, Bedalov A. SIRT1 is dispensable for function of hematopoietic stem cells in adult mice. Blood. 2012;119:1856–1860. doi: 10.1182/blood-2011-09-377077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rimmele P, Lofek-Czubek S, Ghaffari S. Resveratrol increases the bone marrow hematopoietic stem and progenitor cell capacity. Am J Hematol. 2014;89:E235–E238. doi: 10.1002/ajh.23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang QS, Deater M, Schubert K, Marquez-Loza L, Pelz C, Sinclair DA, Grompe M. The Sirt1 activator SRT3025 expands hematopoietic stem and progenitor cells and improves hematopoiesis in fanconi anemia mice. Stem Cell Res. 2015;15:130–140. doi: 10.1016/j.scr.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A, Clemens TL. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Rajendran GK, Liu N, Ware C, Rubin BP, Gu Y. SirT1 modulates the estrogen-insulin-like growth factor-1 signaling for postnatal development of mammary gland in mice. Breast Cancer Res. 2007;9:R1. doi: 10.1186/bcr1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee DH, Kim TS, Lee D, Lim DS. Mammalian sterile 20 kinase 1 and 2 are important regulators of hematopoietic stem cells in stress condition. Sci Rep. 2018;8:942. doi: 10.1038/s41598-018-19637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deveza L, Ortinau L, Lei K, Park D. Comparative analysis of gene expression identifies distinct molecular signatures of bone marrow- and periosteal-skeletal stem/progenitor cells. PLoS One. 2018;13:e0190909. doi: 10.1371/journal.pone.0190909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goncalves KA, Silberstein L, Li S, Severe N, Hu MG, Yang H, Scadden DT, Hu GF. Angiogenin promotes hematopoietic regeneration by dichotomously regulating quiescence of stem and progenitor cells. Cell. 2016;166:894–906. doi: 10.1016/j.cell.2016.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thrall KD, Mahendra S, Jackson MK, Jackson W 3rd, Farese AM, MacVittie TJ. A comparative dose-response relationship between sexes for mortality and morbidity of radiation-induced lung injury in the rhesus macaque. Health Phys. 2019;116:354–365. doi: 10.1097/HP.0000000000000925. [DOI] [PubMed] [Google Scholar]
- 24.Edin NF, Altaner C, Altanerova V, Ebbesen P, Pettersen EO. Low-dose-rate irradiation for 1 hour induces protection against lethal radiation doses but does not affect life span of DBA/2 mice. Dose Response. 2016;14:1559325816673901. doi: 10.1177/1559325816673901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, Mastick GS, Xu C, Yan W. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci U S A. 2014;111:E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.