Abstract
Thymoma, derived from the epithelial cells of the thymus, is a rare malignant tumour type. Following diagnosis with thymoma, patients generally undergo surgical treatment. However, patients with advanced-stage disease are only candidates for chemotherapy and have poor survival. Therefore, it is urgently required to explore effective chemotherapeutic agents for the treatment of thymoma. In the present study, a Bioinformatics analysis was performed to identify novel drugs for thymoma. Differentially expressed genes (DEGs) in thymoma were obtained by Gene Expression Profiling Interactive Analysis. Subsequently, these genes were processed by Connectivity Map analysis to identify suitable compounds. In addition, Metascape software was used to verify drug and target binding. Molecular docking technology was used to verify drug and target binding. Finally, absorption, distribution, metabolism and excretion parameters in the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform database were used for drug screening and for evaluation of the potential clinical value. In total, 2,447 DEGs, including 2,204 upregulated and 243 downregulated genes, were identified from 118 thymoma patients and 339 normal samples. The top 10 drugs displaying the most significant negative correlations were fulvestrant, hesperetin, zidovudine, hydrocortisone, rolitetracycline, ellipticine, sirolimus, quinisocaine, oestradiol (estradiol) and harmine. The predicted targets of these drugs were then confirmed. The score for the association between estrogen receptor 1 (ESR1) and fulvestrant was 0.99. According to the molecular docking analysis, the total scores for the interaction between ESR1 were 10.26, and those for the interaction between tamoxifen and ESR1 were 6.60. The oral bioavailability (%), drug-likeness and drug half-life for hesperetin were 70.31, 0.27 and 15.78, respectively; those for oestradiol were 53.56, 0.32 and 3.50, respectively; and those for harmine were 56.80, 0.13 and 5.04, respectively. In conclusion, several potential therapeutic drugs for thymoma were identified in the present study. The results suggested that the compounds, including fulvestrant, estradiol, hesperetin and ellipticine, represent the most likely drugs for the treatment of thymoma. Future studies should focus on testing these novel compounds in vitro and in vivo.
Keywords: thymoma, differentially expressed genes, connectivity map, molecular docking
Introduction
Thymoma, which is derived from the epithelial cells of the thymus, is a rare malignant tumour type. At the time of diagnosis, patients are usually between 40 and 60 years of age (1); certain patients present with myasthenia graves or other symptoms, including superior vena cava syndrome, dysphagia, cough or chest pain (2). In general, upon diagnosis with thymoma, patients undergo surgical treatment. However, for patients with stage III and IV disease, the 5-year overall survival rates are 74% and <25%, respectively (3), indicating that those patients require further treatment. Furthermore, even after post-operative radiotherapy, the overall survival rates of patients were not significantly improved (4). Therefore, patients with advanced disease are candidates for chemotherapy. According to certain international guidelines, the first-line combination chemotherapy regimens for thymoma include the following: i) Cisplatin, doxorubicin and cyclophosphamide (CAP), ii) CAP with prednisone, iii) cisplatin, doxorubicin, cyclophosphamide and vincristine, iv) cisplatin and etoposide, v) etoposide, ifosfamide and cisplatin and vi) carboplatin/paclitaxel, while the second-line chemotherapy regimens include etoposide, ifosfamide, pemetrexed, 5-fluorouracil or its analogues, gemcitabine and paclitaxel, separately (5). However, even after chemotherapy, the 10-year disease-free survival rate is only 56% for patients with stage III thymoma and 33% for patients with stage IV disease (6).
Therefore, exploration of effective chemotherapeutic agents to treat thymoma is required. However, most drug development strategies primarily involve determining a novel therapeutic target and then searching for compounds that fit the target. Therefore, agent discovery is an expensive endeavor that frequently involves issues with bioavailability first and toxicity later (7). However, drug repositioning, with the aim of identifying novel applications for existing drugs, has been established as a cost-effective strategy. Over the last decade, burgeoning computer technologies have made structure-based compound screening a prevalent tool in early drug identification (8). Among them, Connectivity Map (CMap), which is based on RNA chip technology, is a useful data resource for studying drug mechanisms and drug reallocation (9). Furthermore, all drugs in the database are ranked according to a score, which is derived from conventional measures, including the Pearson correlation coefficient. For instance, those with a score of 1 are the most strongly positively correlated with the query signature, and those with a score of −1 are most strongly negatively correlated. Consequently, by using the differentially expressed genes (DEGs) between thymoma and normal tissues, CMap is able to identify the affected pathways and small-molecule drugs that may be considered potential therapeutic agents for treating thymoma. Accordingly, the use of RNA chip technology may provide novel ideas for the treatment of thymoma.
In the present study, connectivity mapping was performed based on DEGs identified in a large population of patients with thymoma in The Cancer Genome Atlas (TCGA) and GTEx databases, accompanied by the procurement of prospective candidate drugs for the future treatment of thymoma.
Materials and methods
Screening of DEGs in thymoma
Gene expression profiling interactive analysis (access: August 9, 2018; GEPIA; http://gepia.cancer-pku.cn/) was used to identify the DEGs in thymoma. GEPIA uses data from TCGA and GTEx projects and is a freely available tool to consign customizable functionalities (10). Four-way analysis of variance (ANOVA) was used to calculate differential expression based on variables, including sex, age, ethnicity and disease stage. First, on the website, all the expression profiles were transformed to the log2 [transcripts per million (TPM)+1] scale for further calculations. Simultaneously, the median (tumour-normal) was defined by the log2 [fold change (FC)]. The adjusted q-value was then obtained after multiple adjustments using the Benjamini and Hochberg false discovery rate method for each variable. The cut-off values for abnormally expressed mRNAs were a log2FC >3 and a q-value of <0.05.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, and protein-protein interaction (PPI) network of the DEGs
GO and KEGG analyses were used to further understand the roles of DEGs in thymoma (11). The Database for Annotation, Visualization and Integrated Discovery (DAVID) 3.8 (access time: April 10, 2019, http://david.ncifcrf.gov) was applied to identify the GO terms and KEGG pathways in which the DEGs were enriched (12–14). A P-value of <0.05 was considered to indicate significance. Simultaneously, the Search Tool for the Retrieval of Interacting Genes/proteins (STRING) database (access time: April 15, 2019, http://string-db.org), with the cut-off criterion of a combined score of >0.9, was utilized to provide an appraisal and integration of the PPI network (15,16).
Discovery of potential therapeutic drugs depended on the CMap database
CMap was utilized to identify potential therapeutic drugs for thymoma treatment (17). First, Affymetrix gene chips, one of the most frequently used platforms for expression profiling, were used to convert a gene symbol to a signature in HG-U133A format (18). However, with the limitations of the website, a log2FC >4.0 and a q-value of <0.05 were selected as cut-off values for overexpressed genes. Cutoff values for the downregulated expression of mRNAs were a log2FC of >3.0 and a q-value of <0.05. After the data were inputted into CMap (access time: April 6, 2018, http://portals.broadinstitute.org/cmap/), the corresponding compounds with scores of <-0.75 were considered potential drugs for the treatment of thymoma. Finally, the top 10 compounds were subjected to further analysis, and their molecular structures were obtained from PubChem (access time: April 6, 2019, http://www.ncbi.nlm.nih.gov/pccompound) (19).
Construction of the drug-target network
A drug-target network was constructed to further examine the potential mechanisms of action of the compounds. The freely available STITCH software (access time: April 11, 2019, http://prion.bchs.uh.edu/stitch/) was used to construct the drug-target network and to provide genetic identification (20). A higher score indicated a greater likelihood that the drug targeted the tested gene product (21).
Molecular docking analysis of the interactions between proteins and compounds
The Surflex-Dock program in Sybyl version X-2.0 was used to verify the interactions between drugs and targets. The program obtained particular information on how these novel drugs exert their anti-tumour activity. Simultaneously, the docking scores and crash were calculated to represent binding affinities and the degree of inappropriate penetration into the protein by the ligand (22–24).
Functional annotation of the targets
To further explore the functions of the compounds in combination with the target, KEGG pathway and GO term analyses of the targets were performed using the tools mentioned above.
Screening compounds based on absorption, distribution, metabolism and excretion (ADME) parameters in the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) database
The ADME parameters were used for drug screening and evaluation to further determine the potential clinical value of the compounds. The TCMSP database (access time: April 6, 2018, http://sm.nwsuaf.edu.cn/lsp/tcmsp.php) was used to query the ADME parameters of the top 10 drugs identified above (25). The compounds were then screened with the following criteria: Oral bioavailability (OB) ≥30%, drug-likeness (DL) ≥0.18 and drug half-life (HL) ≥4 h.
Results
GO and pathway analyses of the DEGs associated with thymoma
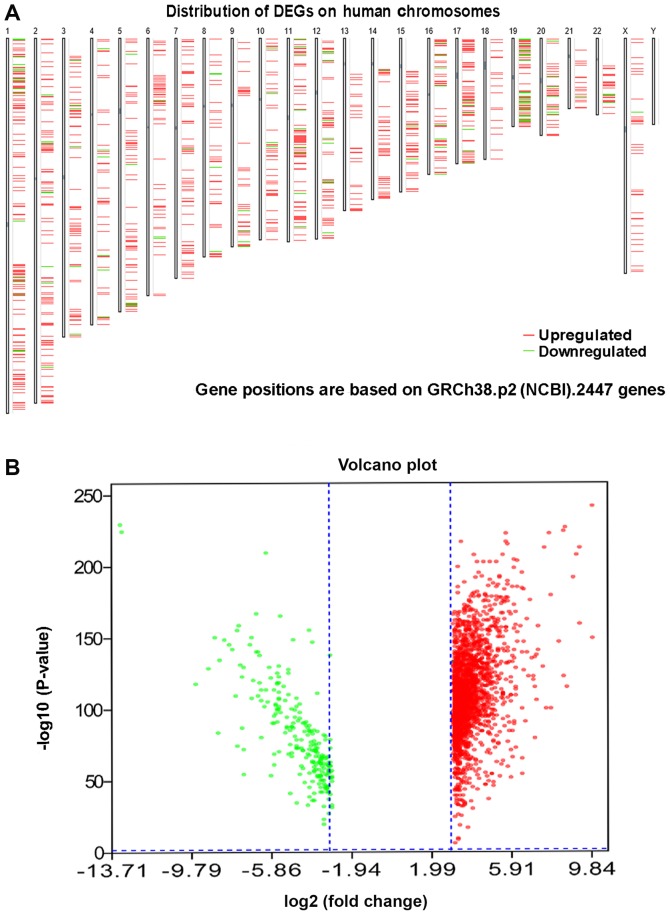
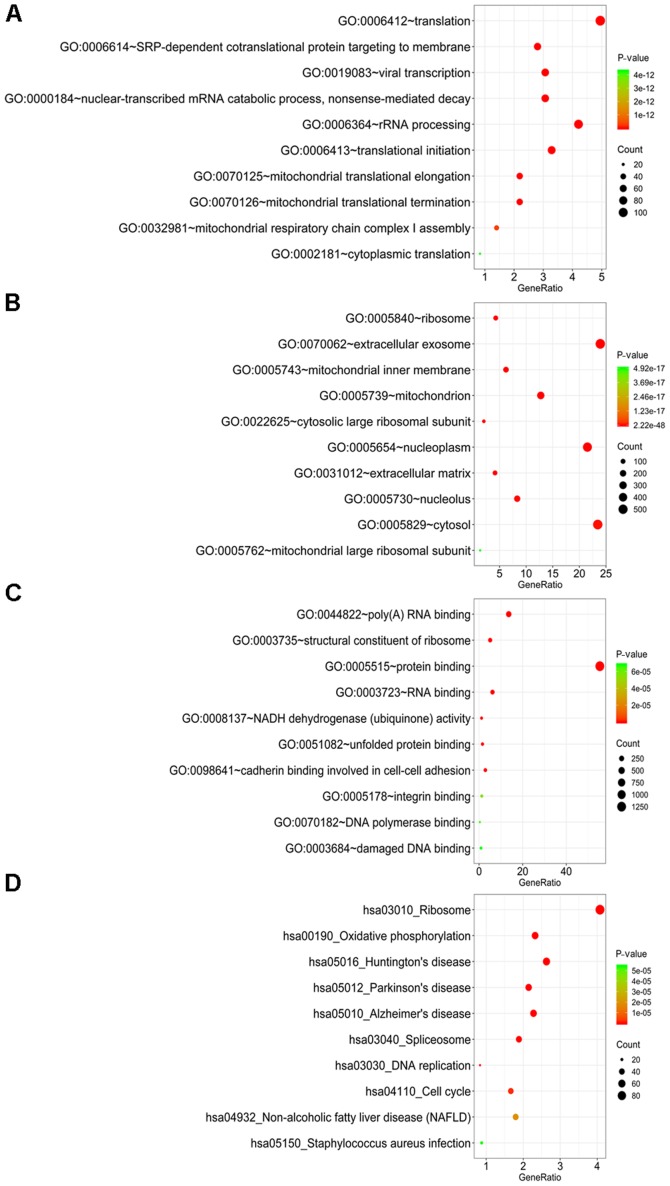
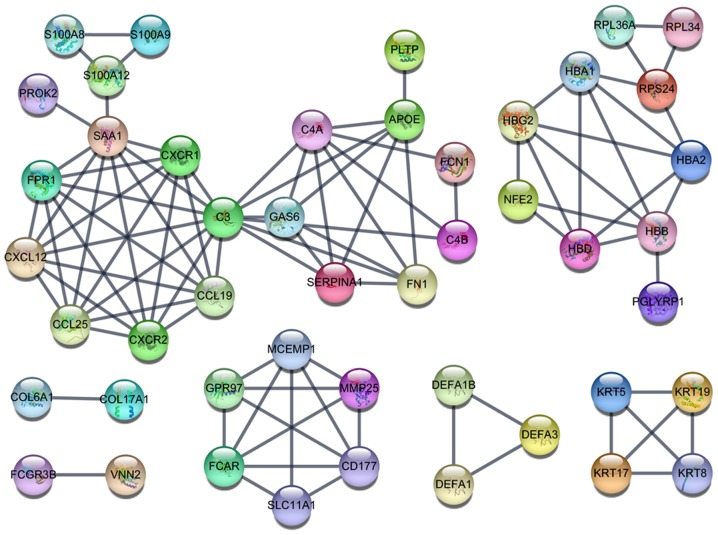
In total, 2,447 DEGs, including 2,204 upregulated and 243 downregulated genes, were identified in 118 thymoma patients and 339 normal samples (2 normal tissues and 337 normal blood samples) (Fig. 1; Table I). A GO analysis was further employed to determine the possible molecular mechanisms of the DEGs. In the category biological process (BP), these genes were most enriched in ‘translation’, ‘SRP-dependent co-translational protein targeting to membrane’, ‘viral transcription’, ‘nuclear-transcribed mRNA catabolic process, nonsense-mediated decay’ and ‘rRNA processing’ (Fig. 2A). In the category cellular component (CC), ‘ribosome’, ‘extracellular exosome’, ‘mitochondrial inner membrane’, ‘mitochondrion’ and ‘cytosolic large ribosomal subunit’ were the most prominent terms (Fig. 2B). In the category molecular function (MF), these genes were significantly involved in ‘poly(A) RNA binding’, ‘structural constituent of ribosome’, ‘protein binding’, ‘RNA binding’ and ‘NADH dehydrogenase (ubiquinone) activity’ (Fig. 2C). In the KEGG pathway analysis, ‘ribosome’, ‘oxidative phosphorylation’, ‘Huntington's disease’, ‘Parkinson's disease’ and ‘Alzheimer's disease’ were the most prominent pathways (Fig. 2D). Among these pathways, ‘ribosome’, with 93 genes included, was the most significant pathway. The PPI networks comprising the DEGs revealed that certain genes, including complement C3 (C3), Serum amyloid A1 (SAA1), C-X-C motif chemokine receptor 1 (CXCR1), C-X-C motif chemokine receptor 2 (CXCR2), C-X-C motif chemokine ligand 12 (CXCL12), C-C motif chemokine ligand 19 (CCL19), C-C motif chemokine ligand 25 (CCL25) and Formyl peptide receptor were closely linked with a high degree (Fig. 3).
Figure 1.
DEGs between thymoma and normal tissues. (A) Locations of the DEGs based on GRCh38.p2 (NCBI). (B) Volcano plot displaying the DEGs. DEG, differentially expressed gene; NCBI, National Center for Biotechnology Information.
Table I.
Clinicopathological features of the thymoma patients (n=118).
| Clinicopathological feature | Number |
|---|---|
| Age (years) | |
| <40 | 10 |
| 40–60 | 51 |
| >60 | 57 |
| Sex | |
| Male | 62 |
| Female | 56 |
| Ethnicity | |
| White | 98 |
| Black | 6 |
| Asian | 12 |
| Not specified | 2 |
| Stage | |
| I | 35 |
| II | 60 |
| III | 15 |
| IV | 6 |
| Not available | 2 |
Figure 2.
GO terms and KEGG pathway analysis for the differentially expressed genes. (A-C) GO terms in the categories (A) biological process, (B) molecular function and (C) cellular component; (D) KEGG pathway analysis. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens.
Figure 3.
The PPI network of the significant DEGs. The PPI network of DEGs with the cut-off criterion of a combined score >0.9 had a total of 47 nodes and 96 edges. PPI, protein-protein interaction; DEG, differentially expressed gene.
Potential therapeutic drugs identified from the CMap database
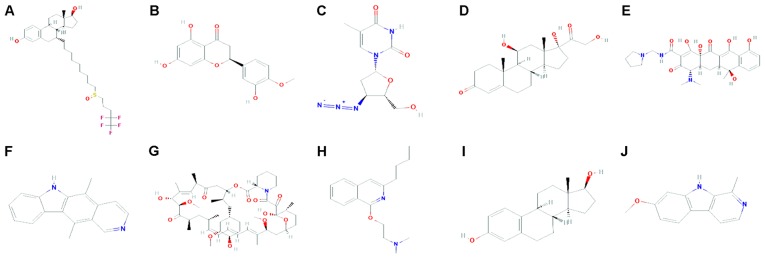
CMap analysis identified 5,000 compounds correlated with the DEGs. According to the score rankings, 769 drugs scored <-0.75 and were considered potential thymoma therapeutics. The top 10 compounds displaying the strongest negative correlation were fulvestrant, hesperetin, zidovudine, hydrocortisone, rolitetracycline, ellipticine, sirolimus, quinisocaine, oestradiol and harmine (Fig. 4; Table II).
Figure 4.
Molecular structures of the top ten drugs. (A) Fulvestrant; (B) hesperetin; (C) zidovudine; (D) hydrocortisone; (E) rolitetracycline; (F) ellipticine; (G) sirolimus; (H) quinisocaine; (I) estradiol; (J) harmine.
Table II.
Connectivity map results for the 10 strongest negative correlation compounds according to CMap.
| Drug name | Dose | Connectivity score | Up score | Down score |
|---|---|---|---|---|
| Fulvestrant | 1 µM | −1.00 | −0.25 | 0.38 |
| Hesperetin | 13 µM | −0.99 | −0.25 | 0.37 |
| Zidovudine | 15 µM | −0.99 | −0.25 | 0.37 |
| Hydrocortisone | 11 µM | −0.99 | −0.24 | 0.38 |
| Rolitetracycline | 8 µM | −0.98 | −0.23 | 0.38 |
| Ellipticine | 16 µM | −0.98 | −0.22 | 0.40 |
| Sirolimus | 100 nM | −0.98 | −0.23 | 0.39 |
| Quinisocaine | 13 µM | −0.98 | −0.23 | 0.38 |
| Estradiol | 100 nM | −0.97 | −0.24 | 0.37 |
| Harmine | 16 µM | −0.97 | −0.21 | 0.40 |
All data are for the MCF7 cell line. Up score and down score represent induced and repressed of the expression in the list respectively. The connectivity score combines the up score and the down score.
Construction of the drug-target network
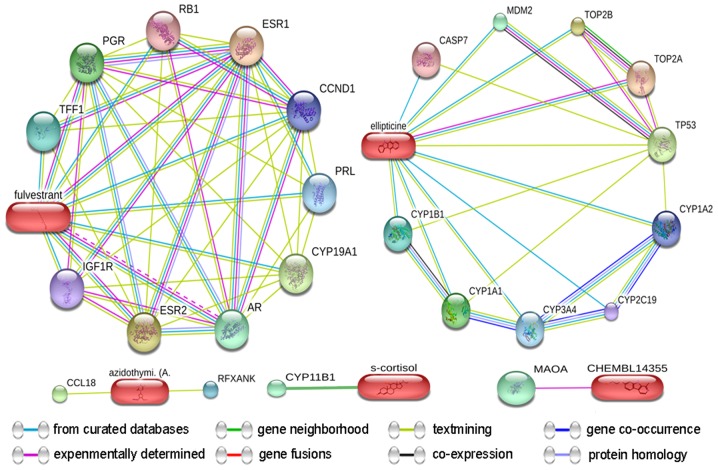
The targets of the test drugs were identified from the STITCH database (20) (Fig. 5). The predicted targets of the top 10 drugs predicted for thymoma were confirmed, and the predicted scores for the interactions of DNA topoisomerase IIα (TOP2A), TOP2B and tumour protein 53 with ellipticine were 0.93, 0.86 and 0.84, respectively. Similarly, the scores for the interactions of estrogen receptor 1 (ESR1), ESR2 and cytochrome P450 family 19 subfamily A member 1 (CYP19A1) with fulvestrant were 0.99, 0.99 and 0.99, respectively. The score for the interaction of CYP11B1 and s-cortisol (hydrocortisone) was 0.90. Furthermore, the scores for the interactions between C-C motif chemokine ligand 18 and regulatory factor X-associated ankyrin-containing protein with azidothymidine (zidovudine) were 0.44 and 0.40, respectively. The score for the interaction of monoamine oxidase A with CHEMBL14355 (harmine) was 0.52 (Table III).
Figure 5.
Drug-target networks of five compounds curated from the STITCH database. Pill-shaped and spheres nodes represent the compounds and proteins respectively.
Table III.
Predicted targets of the drugs identified from the STITCH database.
| Drug/targets | Combined score |
|---|---|
| Ellipticine | |
| TOP2A | 0.93 |
| TOP2B | 0.86 |
| TP53 | 0.84 |
| CYP1A1 | 0.78 |
| MDM2 | 0.75 |
| CYP1B1 | 0.74 |
| CYP3A4 | 0.73 |
| CYP1A2 | 0.73 |
| CASP7 | 0.70 |
| CYP2C19 | 0.70 |
| Fulvestrant | |
| ESR1 | 1.00 |
| ESR2 | 0.99 |
| CYP19A1 | 0.99 |
| AR | 0.97 |
| PGR | 0.97 |
| TFF1 | 0.95 |
| PRL | 0.94 |
| CCND1 | 0.92 |
| IGF1R | 0.92 |
| RB1 | 0.91 |
| S-cortisol | |
| CYP11B1 | 0.90 |
| Azidothymidine (zidovudine) | |
| CCL18 | 0.44 |
| RFXANK | 0.40 |
| CHEMBL14355 | |
| MAOA | 0.52 |
TOP2A, DNA topoisomerase II alpha; TP53, Tumor protein P53; CYP1A1, Cytochrome P450 family 1 subfamily a member 1; MDM2, MDM2 proto-oncogene; CASP7, Caspase 7; ESR1, Estrogen receptor 1; AR, Androgen receptor; PGR, Progesterone receptor; TFF1, Trefoil factor 1; PRL, Prolactin; CCND1, Cyclin D1; IGF1R, Insulin like growth factor 1 receptor; RB1, RB transcriptional corepressor 1; CCL18, C-C motif chemokine ligand 18; RFXANK, Regulatory factor X associated ankyrin containing protein; MAOA, Monoamine oxidase A.
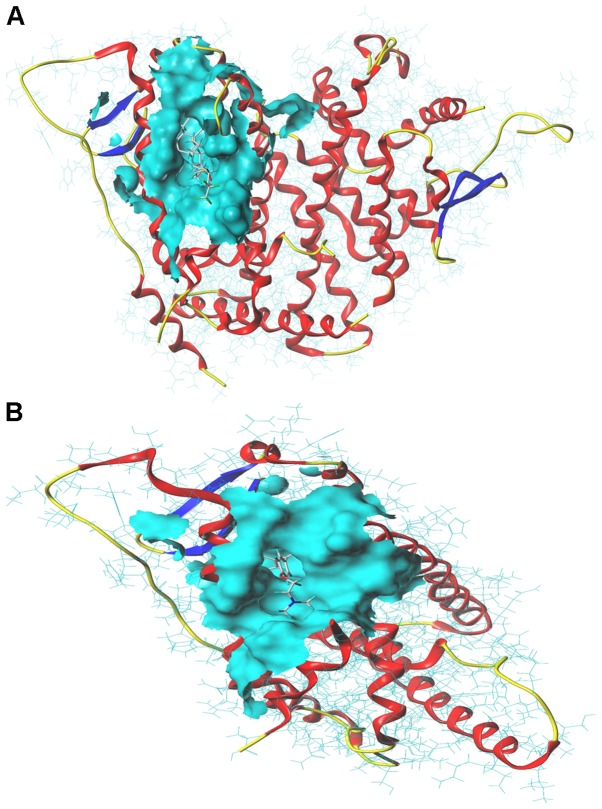
Molecular docking analysis of the interactions between proteins and compounds
Molecular docking analysis was performed to confirm the interactions between drugs and protein targets. The total scores, crash and polar for the fulvestrant and ESR1 interaction were 10.26, −3.72 and 2.08 respectively (Fig. 6A). The total scores, crash and polar for the tamoxifen and ESR1 interaction were 6.60, −4.02 and 0, respectively (Fig. 6B).
Figure 6.
Molecular docking test for compounds and targets. (A) Interaction between fulvestrant and ESR1. (B) Interaction between tamoxifen and ESR1. ESR1, estrogen receptor 1.
Functional annotation of the drug targets
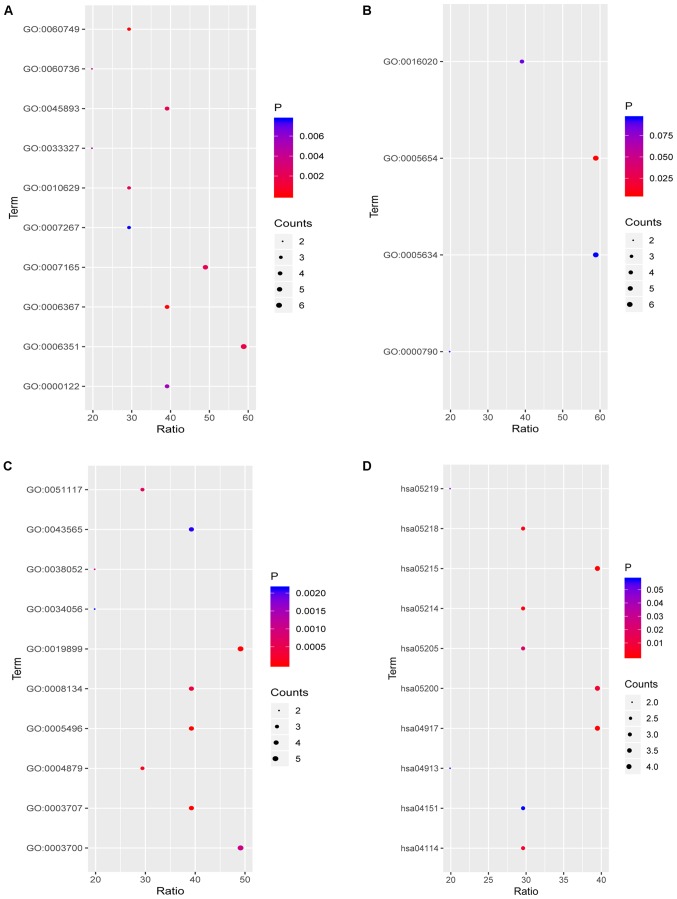
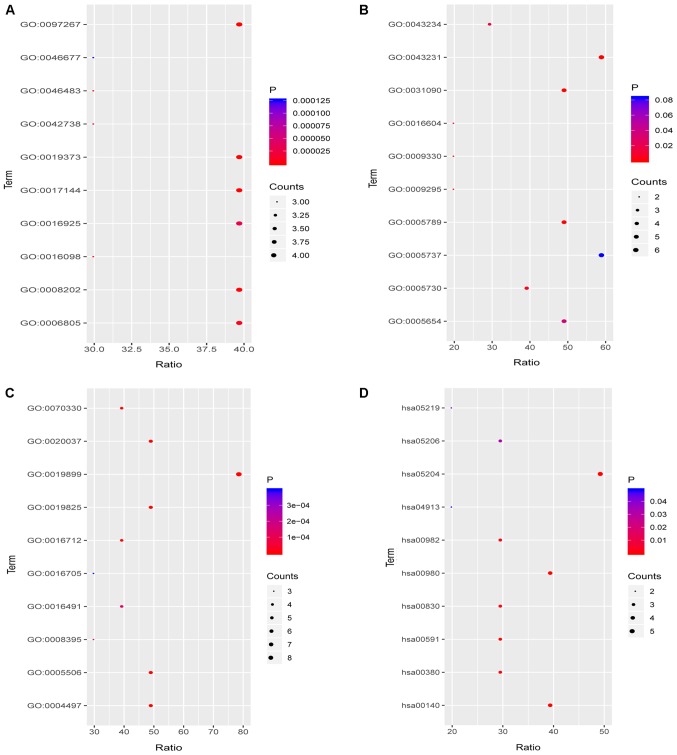
To further explore the functions of the compounds in combination with the target, functional annotation, including GO and KEGG pathway analyses, were performed. The results revealed that the targets for fulvestrant in the category BP were significantly involved in ‘mammary gland alveolus development’, ‘transcription initiation from RNA polymerase II promoter’, ‘transcription, DNA-templated’, ‘positive regulation of transcription, DNA-templated’ and ‘signal transduction’ (Fig. 7A). However, in the CC category, the targets were only significantly involved in ‘nucleoplasm’ (Fig. 7B). In the category MF, the targets were mainly enriched in ‘steroid binding’, ‘steroid hormone receptor activity’, ‘enzyme binding’, ‘RNA polymerase II transcription factor activity, ligand’ and ‘transcription factor binding’ (Fig. 7C). Finally, in the KEGG pathway analysis of fulvestrant, ‘prolactin signaling pathway’, ‘prostate cancer’, ‘glioma’, ‘melanoma’ and ‘oocyte meiosis’ were the most prominent pathways (Fig. 7D; Table IV). Target genes of ellipticine in the category BP were mainly enriched in ‘omega-hydroxylase P450 pathway’, ‘epoxygenase P450 pathway’, ‘drug metabolic process’, ‘steroid metabolic process’ and ‘monoterpenoid metabolic process’ (Fig. 8A). In the CC category, the targets were mainly enriched in ‘organelle membrane’, ‘intracellular membrane-bounded organelle’, ‘endoplasmic reticulum membrane’, ‘nucleoid’ and ‘DNA topoisomerase complex (ATP-hydrolyzing)’ (Fig. 8B). Regarding the enrichment of targets of ellipticine in the category MF, they were significantly involved in ‘enzyme binding’, ‘oxygen binding’, ‘monooxygenase activity’, ‘oxidoreductase activity’ (acting on paired donors with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor and the incorporation of one atom of oxygen) and ‘aromatase activity’ (Fig. 8C). In the KEGG pathway analysis of ellipticine, ‘chemical carcinogenesis’, ‘steroid hormone biosynthesis’, ‘metabolism of xenobiotics by cytochrome P450’, ‘linoleic acid metabolism’ and ‘tryptophan metabolism’ were the most prominent pathways (Fig. 8D; Table V).
Figure 7.
Functional annotation of the targets of fulvestrant. (A-C) GO terms in the categories (A) biological process, (B) molecular function and (C) cellular component; (D) KEGG pathway analysis. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens.
Table IV.
Functional annotation of the targets of fulvestrant.
| ID | Description | Category/type | Gene symbol |
|---|---|---|---|
| GO:0060749 | Mammary gland alveolus development | Biological_process | AR, CCND1, ESR1 |
| GO:0006367 | Transcription initiation from RNA polymerase II promoter | Biological_process | PGR, AR, ESR1, ESR2 |
| GO:0006351 | Transcription, DNA-templated | Biological_process | PGR, AR, CCND1, ESR1, RB1, ESR2 |
| GO:0045893 | Positive regulation of transcription, DNA-template | Biological_process | AR, ESR1, RB1, ESR2 |
| GO:0007165 | Signal transduction | Biological_process | PGR, IGF1R, AR, ESR1, ESR2 |
| GO:0010629 | Negative regulation of gene expression | Biological_process | PGR, ESR1, RB1 |
| GO:0060736 | Prostate gland growth | Biological_process | AR, CYP1A1 |
| GO:0033327 | Leydig cell differentiation | Biological_process | AR, CCND1 |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | Biological_process | CCND1, ESR1, RB1, ESR2 |
| GO:0007267 | Cell-cell signaling | Biological_process | PGR, AR, ESR2 |
| GO:0005654 | Nucleoplasm | Cellular_component | PGR, AR, CCND1, ESR1, RB1, ESR2 |
| GO:0016020 | Membrane | Cellular_component | IGF1R, CCND1, ESR1, CYP19A1 |
| GO:0000790 | Nuclear chromatin | Cellular_component | AR, ESR1 |
| GO:0005634 | Nucleus | Cellular_component | PGR, AR, CCND1, ESR1, RB1, ESR2 |
| GO:0005496 | Steroid binding | Molecular_function | PGR, AR, ESR1, ESR2 |
| GO:0003707 | Steroid hormone receptor activity | Molecular_function | PGR, AR, ESR1, ESR2 |
| GO:0019899 | Enzyme binding | Molecular_function | PGR, AR, CCND1, ESR1, ESR2 |
| GO:0004879 | RNA polymerase II transcription factor activity, ligand | Molecular_function | AR, ESR1, ESR2 |
| GO:0008134 | Transcription factor binding | Molecular_function | AR, CCND1, ESR1, RB1 |
| GO:0051117 | ATPase binding | Molecular_function | PGR, AR, ESR1 |
| GO:0003700 | Transcription factor activity, sequence | Molecular_function | PGR, AR, ESR1, RB1, ESR2 |
| GO:0038052 | RNA polymerase II transcription factor activity, estrogen | Molecular_function | ESR1, ESR2 |
| GO:0043565 | Sequence-specific DNA binding | Molecular_function | PGR, AR, ESR1, ESR2 |
| GO:0034056 | Estrogen response element binding | Molecular_function | ESR1, ESR2 |
| hsa04917 | Prolactin signaling pathway | KEGG | CCND1, ESR1, ESR2, PRL |
| hsa05215 | Prostate cancer | KEGG | IGF1R, AR, CCND1, RB1 |
| hsa05214 | Glioma | KEGG | IGF1R, CCND1, RB1 |
| hsa05218 | Melanoma | KEGG | IGF1R, CCND1, RB1 |
| hsa04114 | Oocyte meiosis | KEGG | PGR, IGF1R, AR |
| hsa05200 | Pathways in cancer | KEGG | IGF1R, AR, CCND1, RB1 |
| hsa05205 | Proteoglycans in cancer | KEGG | IGF1R, CCND1, ESR1 |
| hsa05219 | Bladder cancer | KEGG | CCND1, RB1 |
| hsa04913 | Ovarian steroidogenesis | KEGG | IGF1R, CYP19A1 |
| hsa04151 | PI3K/Akt signaling pathway | KEGG | IGF1R, CCND1, PRL |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens; AR, Androgen receptor; CCND1, Cyclin D1; ESR1, Estrogen receptor 1; PGR, Progesterone receptor; RB1, RB transcriptional corepressor 1; IGF1R, Insulin like growth factor 1 receptor; CYP1A1, Cytochrome P450 family 1 subfamily a member 1; PRL, Prolactin.
Figure 8.
Functional annotation of the targets of ellipticine. (A-C) GO terms in the categories (A) biological process, (B) molecular function and (C) cellular component; (D) KEGG pathway analysis. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens.
Table V.
Functional annotation of the targets of ellipticine.
| ID | Description | Type | Genes |
|---|---|---|---|
| GO:0097267 | Omega-hydroxylase P450 pathway | Biological_process | CYP1B1, CYP1A1, CYP2C19, CYP1A2 |
| GO:0019373 | Epoxygenase P450 pathway | Biological_process | CYP1B1, CYP1A1, CYP2C19, CYP1A2 |
| GO:0017144 | Drug metabolic process | Biological_process | CYP3A4, CYP1A1, CYP2C19, CYP1A2 |
| GO:0008202 | Steroid metabolic process | Biological_process | CYP3A4, CYP1B1, CYP1A1, CYP2C19 |
| GO:0016098 | Monoterpenoid metabolic process | Biological_process | CYP3A4, CYP2C19, CYP1A2 |
| GO:0046483 | Heterocycle metabolic process | Biological_process | CYP3A4, CYP2C19, CYP1A2 |
| GO:0006805 | Xenobiotic metabolic process | Biological_process | CYP3A4, CYP1B1, CYP2C19, CYP1A2 |
| GO:0042738 | Exogenous drug catabolic process | Biological_process | CYP3A4, CYP2C19, CYP1A2 |
| GO:0016925 | Protein sumoylation | Biological_process | TP53, MDM2, TOP2B, TOP2A |
| GO:0046677 | Response to antibiotic | Biological_process | CYP1A1, TP53, MDM2 |
| GO:0031090 | Organelle membrane | Cellular_component | CYP3A4, CYP1B1, CYP1A1, CYP2C19, CYP1A2 |
| GO:0043231 | Intracellular membrane-bound organelle | Cellular_component | CYP3A4, CYP1B1, CYP1A1, CYP2C19, CASP7, CYP1A2 |
| GO:0005789 | Endoplasmic reticulum membrane | Cellular_component | CYP3A4, CYP1B1, CYP1A1, CYP2C19, CYP1A2 |
| GO:0009295 | Nucleoid | Cellular_component | TOP2B, TOP2A |
| GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing) | Cellular_component | TOP2B, TOP2A |
| GO:0005730 | Nucleolus | Cellular_component | TP53, MDM2, TOP2B, TOP2A |
| GO:0016604 | Nuclear body | Cellular_component | TP53, MDM2 |
| GO:0043234 | Protein complex | Cellular_component | TP53, MDM2, TOP2A |
| GO:0005654 | Nucleoplasm | Cellular_component | CASP7, TP53, MDM2, TOP2B, TOP2A |
| GO:0005737 | Cytoplasm | Cellular_component | CYP3A4, CASP7, TP53, MDM2, TOP2B, TOP2A |
| GO:0019899 | Enzyme binding | Molecular_function | CYP3A4, CYP1A1, CYP2C19, TP53, MDM2, CYP1A2, TOP2B, TOP2A |
| GO:0019825 | Oxygen binding | Molecular_function | CYP3A4, CYP1B1, CYP1A1, CYP2C19, CYP1A2 |
| GO:0004497 | Monooxygenase activity | Molecular_function | CYP3A4, CYP1B1, CYP1A1, CYP2C19, CYP1A2 |
| GO:0016712 | Reduced flavin or flavoprotein as one donor | Molecular_function | CYP3A4, CYP1B1, CYP1A1, CYP1A2 |
| GO:0070330 | Aromatase activity | Molecular_function | CYP3A4, CYP1B1, CYP1A1, CYP1A2 |
| GO:0020037 | Heme binding | Molecular_function | CYP3A4, CYP1B1, CYP1A1, CYP2C19, CYP1A2 |
| GO:0005506 | Iron ion binding | Molecular_function | CYP3A4, CYP1B1, CYP1A1, CYP2C19, CYP1A2 |
| GO:0008395 | Steroid hydroxylase activity | Molecular_function | CYP3A4, CYP1A1, CYP2C19 |
| GO:0016491 | Oxidoreductase activity | Molecular_function | CYP3A4, CYP1A1, CYP2C19, CYP1A2 |
| GO:0016705 | Incorporation or reduction of molecular oxygen | Molecular_function | CYP3A4, CYP1B1, CYP2C19 |
| hsa05204 | Chemical carcinogenesis | KEGG | CYP3A4, CYP1B1, CYP1A1, CYP2C19, CYP1A2 |
| hsa00140 | Steroid hormone biosynthesis | KEGG | CYP3A4, CYP1B1, CYP1A1, CYP1A2 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | KEGG | CYP3A4, CYP1B1, CYP1A1, CYP1A2 |
| hsa00591 | Linoleic acid metabolism | KEGG | CYP3A4, CYP2C19, CYP1A2 |
| hsa00380 | Tryptophan metabolism | KEGG | CYP1B1, CYP1A1, CYP1A2 |
| hsa00830 | Retinol metabolism | KEGG | CYP3A4, CYP1A1, CYP1A2 |
| hsa00982 | Drug metabolism-cytochrome P450 | KEGG | CYP3A4, CYP2C19, CYP1A2 |
| hsa05206 | MicroRNAs in cancer | KEGG | CYP1B1, TP53, MDM2 |
| hsa05219 | Bladder cancer | KEGG | TP53, MDM2 |
| hsa04913 | Ovarian steroidogenesis | KEGG | CYP1B1, CYP1A1 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens; CYP1A1, Cytochrome P450 family 1 subfamily a member 1; TP53, Tumor protein P53; MDM2, MDM2 proto-oncogene; TOP2B, DNA topoisomerase II beta; CASP7, Caspase 7.
Screening of compounds based on ADME parameters in the TCMSP database
Hesperetin, oestradiol and harmine were searched in the TCMSP database. The OB (%), DL and HL for hesperetin were 70.31, 0.27 and 15.78, respectively; those for oestradiol were 53.56, 0.32 and 3.50, respectively; and those for harmine were 56.80, 0.13 and 5.04, respectively (Table VI).
Table VI.
Absorption, distribution, metabolism and excretion parameters for three drugs from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform database.
| Drug name | Oral bioavailability (%) | Drug-likeness (0–1) | Drug half-life (h) |
|---|---|---|---|
| Hesperetin | 70.31 | 0.27 | 15.78 |
| Estradiol | 53.56 | 0.32 | 3.5 |
| Harmine | 56.8 | 0.13 | 5.04 |
Discussion
In the present study, it was speculated that the potential therapeutic drugs for thymoma are compounds that are matched with DEGs known to be associated with the occurrence and development of tumours. First, by using the DEG data from the GEPIA database, correlations between the genes and previously known pharmaceutical compounds were revealed in CMap. Subsequently, the top 10 molecules with the lowest negative correlations were obtained and they were considered as potential therapeutic drugs for further analysis. In addition, a drug-target network was constructed to examine the potential mechanisms of action of the compounds. Molecular docking analysis was then performed to confirm the interactions between the drugs and protein targets. Furthermore, the ADME parameters were inquired to determine the potential clinical value of the compounds.
According to the GEPIA tool, 2,447 DEGs were identified in 118 thymoma patients and 339 normal samples. After functional annotation analysis, it was determined that the DEGs were enriched in ‘ribosome’, ‘oxidative phosphorylation’, ‘spliceosome’, ‘DNA replication’ and ‘cell cycle’, which indicated that the DEGs may affect cell growth and have an important role in the occurrence and development of thymoma. Then, based on the DEGs, potential therapeutic drugs were identified using the CMap database, including fulvestrant, hesperetin, zidovudine, hydrocortisone, rolitetracycline, ellipticine, sirolimus, quinisocaine, oestradiol and harmine. Then, to examine the effect of the potential drugs, a drug-target network was constructed and a molecular docking analysis was performed. Compared with the docking score of tamoxifen and ESR1, the score of fulvestrant and ESR1 was high, which indicates a high interaction between fulvestrant and ESR1. Among these compounds, tamoxifen, as an ESR1 inhibitor, remains the first-line of medication for the treatment of ESR1+ breast cancer (26). In addition, the ESR1 protein localizes to the nucleus and has been proven important in the pathological processes of several cancer types, including breast and endometrial cancers (27,28). Furthermore, functional enrichment analysis indicated that the targets of fulvestrant significantly accumulated in ‘prolactin signaling pathway’, ‘prostate cancer’, ‘glioma’ and ‘melanoma’. The results indicated that fulvestrant may combine with ESR1 to affect the treatment of thymoma. Finally, using the TCMSP database, it was determined that the compounds oestrogen, oestradiol and harmine may potentially be of high clinical value. In summary, the compounds may have an important role in the treatment of thymoma. Among these compounds, fulvestrant has been used to treat breast and prostate cancers. Regardless of endocrine tolerance or the levels of hormone receptor expression, fulvestrant significantly improves the survival of patients with non-progressive breast cancer (29–31). In addition, fulvestrant also represents a treatment option for patients with recurrent hormone receptor-positive or HER2-negative metastatic breast cancer (32).
Hesperetin is a bioflavonoid from citrus fruit. Based on experimental evidence, hesperetin possesses anti-oxidant and free radical-quenching activities. This compound also induces apoptotic cell death. In addition, hesperetin reportedly exerts an anti-cancer effect on various cancer cell lines, including breast cancer, prostate cancer, human colon adenocarcinoma and hepatocellular carcinoma cells (33–37). Based on ADME values of the compound molecules from the TCMSP database, it was revealed that hesperetin, estradiol and harmine may have good prospects. Among them, hesperetin caught our attention. The consequence of it for oral bioavailability was 70.31. A high OB (%) is frequently considered a key factor to judge the drug-like properties of compounds as therapeutic agents. Furthermore, as hesperetin is derived from citrus fruit, it may be easy to produce on a large scale. However, due to its poor water solubility, its clinical use is restricted. Numerous studies have been undertaken to improve the bioavailability of flavonoids (33). In summary, hesperetin may not only be used as a promising compound to treat thymoma in the future, but may also be commonly used in the treatment of other tumour types.
Zidovudine is an inhibitor of HIV replication that may reverse neurological dysfunction induced by HIV and ameliorate certain clinical abnormalities (38,39). Hydrocortisone is the major glucocorticoid and its synthetic counterpart is used to treat inflammation, allergy, shock and certain neoplasms (40). Rolitetracycline is a broad-spectrum antibiotic (41). Sirolimus is a potent immunosuppressant (42). Quinisocaine blocks nerve conduction when applied to nervous tissues at appropriate concentrations.
Estrogen has been consistently reported to affect the advancement of thymoma (43–45). However, the therapeutic value of fulvestrant and hesperetin for thymoma has not been previously reported. Simultaneously, the other compounds among the top 10 have also not been reported to be suitable for the treatment of cancer.
The limitations to the present study include the following: First, the DEGs should be further validated in vitro to determine their specific expression in thymoma. In addition, the exact DEGs between the different groups of patients and at different stages of the disease should be determined in vitro to further identify potential compounds, particularly in patients aged 40–60 years and in stage III/IV. Finally, further research should focus on in vitro and in vivo tests prior to the clinical application of these compounds.
In summary, the development of compounds or combinations of drugs remains a requirement in order to improve chemotherapy. The present study identified compounds that may represent novel treatments for thymoma and may reduce the range of potential drugs for treating thymoma.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- DEG
differentially expressed gene
- GEPIA
gene expression profiling interactive analysis
- CMap
connectivity map
- TCGA
The Cancer Genome Atlas
- GO
Gene Ontology
- PPI
protein-protein interaction
- TCMSP
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform
- ADME
absorption, distribution, metabolism and excretion
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during this study are available from the corresponding author on reasonable request.
Authors' contributions
The study was designed by GC, HY, QL and RL. XW, PL, YL, GC, HY, YH and QL were involved in the statistical analysis. HY, YH, QL and RL were involved in drafting the manuscript and critically revising it for important intellctual content. YH, QL and RL gave final approval for the version of the manuscript to be published.
Each author sufficiently participated in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work to ensure that questions regarding the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Safieddine N, Liu G, Cuningham K, Ming T, Hwang D, Brade A, Bezjak A, Fischer S, Xu W, Azad S, et al. Prognostic factors for cure, recurrence and long-term survival after surgical resection of thymoma. J Thorac Oncol. 2014;9:1018–1022. doi: 10.1097/JTO.0000000000000215. [DOI] [PubMed] [Google Scholar]
- 2.Thomas CR, Wright CD, Loehrer PJ. Thymoma: State of the art. J Clin Oncol. 1999;17:2280–2289. doi: 10.1200/JCO.1999.17.7.2280. [DOI] [PubMed] [Google Scholar]
- 3.Kim DJ, Yang WI, Choi SS, Kim KD, Chung KY. Prognostic and clinical relevance of the World Health Organization schema for the classification of thymic epithelial tumors: A clinicopathologic study of 108 patients and literature review. Chest. 2005;127:755–761. doi: 10.1378/chest.127.3.755. [DOI] [PubMed] [Google Scholar]
- 4.Utsumi T, Shiono H, Kadota Y, Matsumura A, Maeda H, Ohta M, Yoshioka Y, Koizumi M, Inoue T, Okumura M. Postoperative radiation therapy after complete resection of thymoma has little impact on survival. Cancer. 2009;115:5413–5420. doi: 10.1002/cncr.24618. [DOI] [PubMed] [Google Scholar]
- 5.Ettinger DS, Riely GJ, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Govindan R, et al. Thymomas and thymic carcinomas: Clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2013;11:562–576. doi: 10.6004/jnccn.2013.0072. [DOI] [PubMed] [Google Scholar]
- 6.Venuta F, Rendina EA, Pescarmona EO, De Giacomo T, Vegna ML, Fazi P, Flaishman I, Guarino E, Ricci C. Multimodality treatment of thymoma: A prospective study. Ann Thorac Surg. 1997;64:1585–1592. doi: 10.1016/S0003-4975(97)00629-2. [DOI] [PubMed] [Google Scholar]
- 7.Ma DL, Chan DS, Leung CH. Drug repositioning by structure-based virtual screening. Chem Soc Rev. 2013;42:2130–2141. doi: 10.1039/c2cs35357a. [DOI] [PubMed] [Google Scholar]
- 8.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat Rev Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 9.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 10.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He RQ, Qin MJ, Lin P, Luo YH, Ma J, Yang H, Hu XH, Chen G. Prognostic significance of LncRNA PVT1 and its potential target gene network in human cancers: A comprehensive inquiry based upon 21 cancer types and 9,972 cases. Cell Physiol Biochem. 2018;46:591–608. doi: 10.1159/000488627. [DOI] [PubMed] [Google Scholar]
- 12.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni Z, Wang X, Zhang T, Li L, Li J. Comprehensive analysis of differential expression profiles reveals potential biomarkers associated with the cell cycle and regulated by p53 in human small cell lung cancer. Exp Ther Med. 2018;15:3273–3282. doi: 10.3892/etm.2018.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Liang L, Wei DM, Li JJ, Luo DZ, Chen G, Dang YW, Cai XY. Prognostic microRNAs and their potential molecular mechanism in pancreatic cancer: A study based on The Cancer Genome Atlas and bioinformatics investigation. Mol Med Rep. 2018;17:939–951. doi: 10.3892/mmr.2017.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musa A, Ghoraie LS, Zhang SD, Glazko G, Yli-Harja O, Dehmer M, Haibe-Kains B, Emmert-Streib F. A review of connectivity map and computational approaches in pharmacogenomics. Brief Bioinform. 2018;19:506–523. doi: 10.1093/bib/bbw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalma-Weiszhausz DD, Warrington J, Tanimoto EY, Miyada CG. The affymetrix GeneChip platform: An overview. Methods Enzymol. 2006;410:3–28. doi: 10.1016/S0076-6879(06)10001-4. [DOI] [PubMed] [Google Scholar]
- 19.Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43 (Database Issue) 2015:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu D, Vaishampayan PA, Venkateswaran K, Fox GE. STITCH: Algorithm to splice, trim, identify, track, and capture the uniqueness of 16S rRNAs sequence pairs using public or in-house database. Microb Ecol. 2011;61:669–675. doi: 10.1007/s00248-010-9779-2. [DOI] [PubMed] [Google Scholar]
- 22.Homer RW, Swanson J, Jilek RJ, Hurst T, Clark RD. SYBYL line notation (SLN): A single notation to represent chemical structures, queries, reactions, and virtual libraries. J Chem Inf Model. 2008;48:2294–2307. doi: 10.1021/ci7004687. [DOI] [PubMed] [Google Scholar]
- 23.Koelmel JP, Ulmer CZ, Jones CM, Yost RA, Bowden JA. Common cases of improper lipid annotation using high-resolution tandem mass spectrometry data and corresponding limitations in biological interpretation. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:766–770. doi: 10.1016/j.bbalip.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preston S, Jiao Y, Baell JB, Keiser J, Crawford S, Koehler AV, Wang T, Simpson MM, Kaplan RM, Cowley KJ, et al. Screening of the ‘Open Scaffolds’ collection from Compounds Australia identifies a new chemical entity with anthelmintic activities against different developmental stages of the barber's pole worm and other parasitic nematodes. Int J Parasitol Drugs Drug Resist. 2017;7:286–294. doi: 10.1016/j.ijpddr.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wardell SE, Ellis MJ, Alley HM, Eisele K, VanArsdale T, Dann SG, Arndt KT, Primeau T, Griffin E, Shao J, et al. Efficacy of SERD/SERM Hybrid-CDK4/6 inhibitor combinations in models of endocrine therapy-resistant breast cancer. Clin Cancer Res. 2015;21:5121–5130. doi: 10.1158/1078-0432.CCR-15-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fribbens C, O'Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, Cristofanilli M, Andre F, Loi S, Loibl S, et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2016;34:2961–2968. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 28.Lebeau A, Grob T, Holst F, Seyedi-Fazlollahi N, Moch H, Terracciano L, Turzynski A, Choschzick M, Sauter G, Simon R. Oestrogen receptor gene (ESR1) amplification is frequent in endometrial carcinoma and its precursor lesions. J Pathol. 2008;216:151–157. doi: 10.1002/path.2405. [DOI] [PubMed] [Google Scholar]
- 29.Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: Key mechanisms of response and failure. Oncogene. 2010;29:4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 30.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.VanArsdale T, Boshoff C, Arndt KT, Abraham RT. Molecular pathways: Targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin Cancer Res. 2015;21:2905–2910. doi: 10.1158/1078-0432.CCR-14-0816. [DOI] [PubMed] [Google Scholar]
- 32.Cristofanilli M, Turner NC, Bondarenko I, Ro J, Im SA, Masuda N, Colleoni M, DeMichele A, Loi S, Verma S, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): Final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016;17:425–439. doi: 10.1016/S1470-2045(15)00613-0. [DOI] [PubMed] [Google Scholar]
- 33.Mary Lazer L, Sadhasivam B, Palaniyandi K, Muthuswamy T, Ramachandran I, Balakrishnan A, Pathak S, Narayan S, Ramalingam S. Chitosan-based nano-formulation enhances the anticancer efficacy of hesperetin. Int J Biol Macromol. 2018;107:1988–1998. doi: 10.1016/j.ijbiomac.2017.10.064. [DOI] [PubMed] [Google Scholar]
- 34.Choi EJ. Hesperetin induced G1-phase cell cycle arrest in human breast cancer MCF-7 cells: Involvement of CDK4 and p21. Nutr Cancer. 2007;59:115–119. doi: 10.1080/01635580701419030. [DOI] [PubMed] [Google Scholar]
- 35.Sambantham S, Radha M, Paramasivam A, Anandan B, Malathi R, Chandra SR, Jayaraman G. Molecular mechanism underlying hesperetin-induced apoptosis by in silico analysis and in prostate cancer PC-3 cells. Asian Pac J Cancer Prev. 2013;14:4347–4352. doi: 10.7314/APJCP.2013.14.7.4347. [DOI] [PubMed] [Google Scholar]
- 36.Sivagami G, Vinothkumar R, Bernini R, Preethy CP, Riyasdeen A, Akbarsha MA, Menon VP, Nalini N. Role of hesperetin (a natural flavonoid) and its analogue on apoptosis in HT-29 human colon adenocarcinoma cell line-a comparative study. Food Chem Toxicol. 2012;50:660–671. doi: 10.1016/j.fct.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Song J, Wu D, Wang J, Dong W. Hesperetin induces the apoptosis of hepatocellular carcinoma cells via mitochondrial pathway mediated by the increased intracellular reactive oxygen species, ATP and calcium. Med Oncol. 2015;32:101. doi: 10.1007/s12032-015-0516-z. [DOI] [PubMed] [Google Scholar]
- 38.McLeod GX, Hammer SM. Zidovudine: Five years later. Ann Intern Med. 1992;117:487–501. doi: 10.7326/0003-4819-117-6-487. [DOI] [PubMed] [Google Scholar]
- 39.De Clercq E. HIV resistance to reverse transcriptase inhibitors. Biochemical pharmacology. 1994;47:155–169. doi: 10.1016/0006-2952(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 40.Palacios R, Sugawara I. Hydrocortisone abrogates proliferation of T cells in autologous mixed lymphocyte reaction by rendering the interleukin-2 Producer T cells unresponsive to interleukin-1 and unable to synthesize the T-cell growth factor. Scand J Immunol. 1982;15:25–31. doi: 10.1111/j.1365-3083.1982.tb00618.x. [DOI] [PubMed] [Google Scholar]
- 41.Schuttemeyer W, Reiff K. Clinical observations on the broad spectrum antibiotic reverin. Dtsch Med J. 1959;10:90–91. (In German) [PubMed] [Google Scholar]
- 42.Sehgal SN. Sirolimus: Its discovery, biological properties, and mechanism of action. Transplant Proc. 2003;35(Suppl 3):S7–S14. doi: 10.1016/S0041-1345(03)00211-2. [DOI] [PubMed] [Google Scholar]
- 43.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 44.Okasha SA, Ryu S, Do Y, McKallip RJ, Nagarkatti M, Nagarkatti PS. Evidence for estradiol-induced apoptosis and dysregulated T cell maturation in the thymus. Toxicology. 2001;163:49–62. doi: 10.1016/S0300-483X(01)00374-2. [DOI] [PubMed] [Google Scholar]
- 45.Zoller AL, Schnell FJ, Kersh GJ. Murine pregnancy leads to reduced proliferation of maternal thymocytes and decreased thymic emigration. Immunology. 2007;121:207–215. doi: 10.1111/j.1365-2567.2006.02559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during this study are available from the corresponding author on reasonable request.