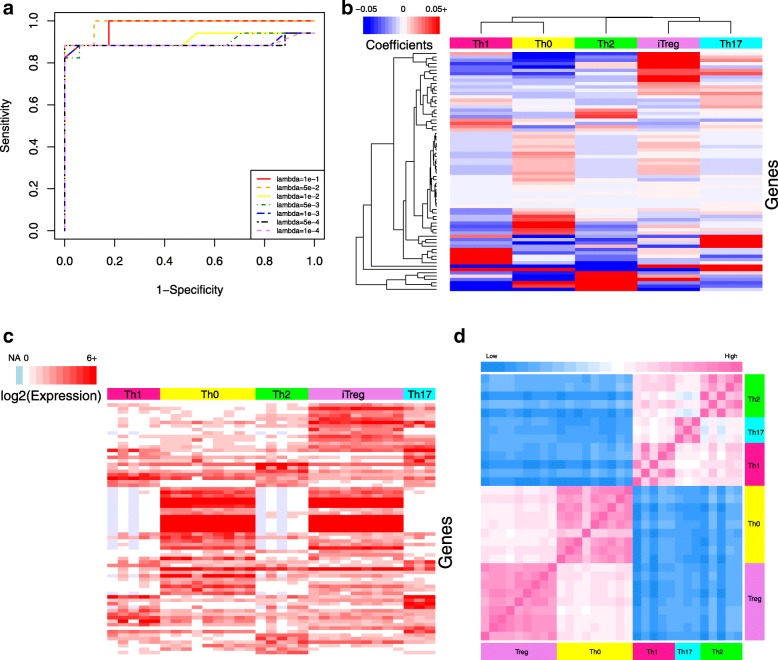
Fig. 5.
Development of T helper cell classifier and similarity heatmaps a ROC curve for the T helper cell classifier was calculated using the indicated lambda values (shown in different colors and line styles) and 10-fold cross validation. The lambda value that maximized the AUC value was used for subsequent calculations. Elastic-net logistic regression to discriminate among five T helper cell types, where the value of the non-zero coefficients (panel b), expression levels (panel c), and similarity map (panel d) for the 72 genes included in the classifier are indicated by color bars for each panel. In panel b, blue to red color scheme indicates coefficients ranging from negative to positive values. Ordering of the genes is the same in panels b and c. In panel c, light blue indicates missing values and the intensity of red color (white/red color scale on the top-left) indicates the log base 2 expression level. A color bar on top of this panel was used to separate samples of each cell type. Panel d illustrates the similarity between samples calculated using an euclidean distance matrix based on the same 72 genes, where the color indicates the distance (pink: high similarity/low distance; blue: low similarity/high distance). Color bar on the top/side of the heatmap indicates the cell type of origin