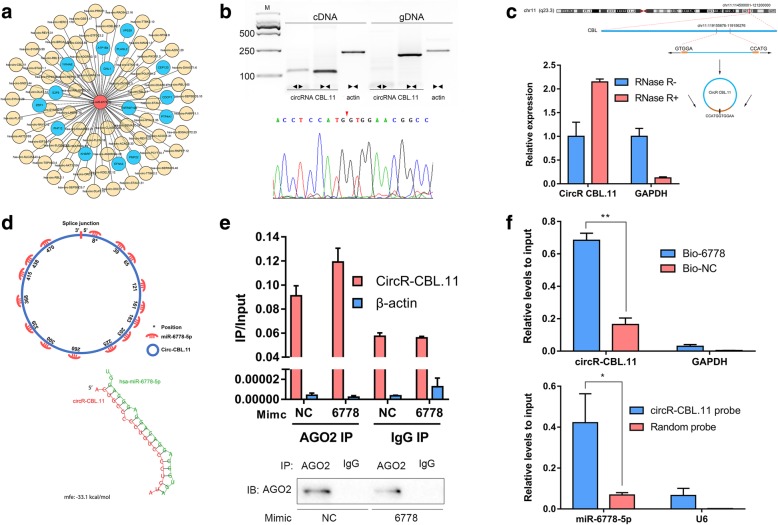
Fig. 5.
CircRNA CBL.11 functions as a competing endogenous RNA by sponging miR-6778-5p. a Cytoscape was used to visualize circRNA-miRNA-mRNA interactions based on the circnet results. In this network, 67 circRNAs (yellow) that interact with miR-6778-5p and the 15 most possible target genes (blue) are presented. b Divergent primers amplify circRNA CBL.11 in cDNA but not genomic DNA (gDNA). Sequencing results of the splice junction are shown in the lower panel. c Schematic diagram of genomic localization and splicing patterns of circRNA CBL.11. Real-time PCR analysis of circRNA CBL.11 and GAPDH after treatment with RNase R. d CircRNA CBL.11 contains 15 binding sites on miR-6778-5p, which was analyzed by RNAhybrid. The binding sites with the smallest minimal free energy are shown in the lower panel. e IP of AGO2 from HCT116 cells transfected with miR-6778-5p or the control mimic. The expression of circRNA CBL.11 was detected by qRT-PCR. β-actin was used as a control. f The biotinylated miR-6778-5p (Bio-6778) or NC control (Bio-NC) was transfected into HCT116 cells. CircRNA CBL.11 was quantified by real-time PCR, after streptavidin capture. GAPDH was used as a control. In the lower panel, miR-6778-5p was pulled down and enriched with a circRNA CBL.11-specific probe and then detected by rear-time PCR