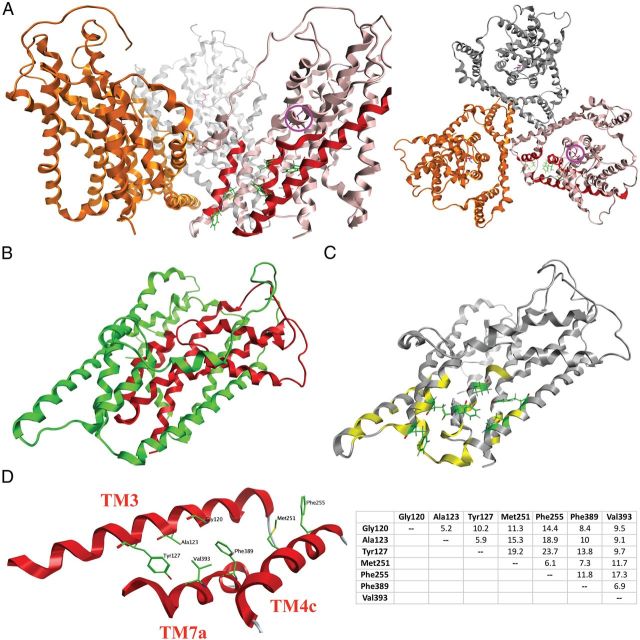
Figure 11.
Homology model of the GLAST monomer. A, The trimeric GLAST complex viewed parallel to the membrane (left) and from the extracellular side of the membrane (right) with the three monomers given in pink, gold, and silver. In the pink monomer, the TM3, TM4c, and TM7a α-helices are highlighted in red, and the seven residues observed to be important for UCPH-101 activity in this study are given as stick representations in green. The localization of the substrate binding site in the pink monomer is indicated by the presence of l-aspartate (in purple, highlighted with a purple circle). B, The chimera N354 monomer. As in Figure 7B, the N354 domains composed of GLAST and GLT-1 regions are given in green and red, respectively. C, The residues in the GLAST monomer subjected to mutagenesis (the specific mutants are given in Table 2). Residues where none of the introduced substitutions affected UCPH-101 activity are given in yellow, whereas the seven residues where selected mutations impaired UCPH-101 potency and/or efficacy are given as stick representations in green. D, The seven residues demonstrated to be important for UCPH-101 activity at GLAST. Left, Detail of the homology model of the GLAST monomer. Spatial orientations of TM3, TM4c, and TM7a and the seven residues. Right, The distances between the seven residues (Cα to Cα distances, in Å).