FIGURE 3.
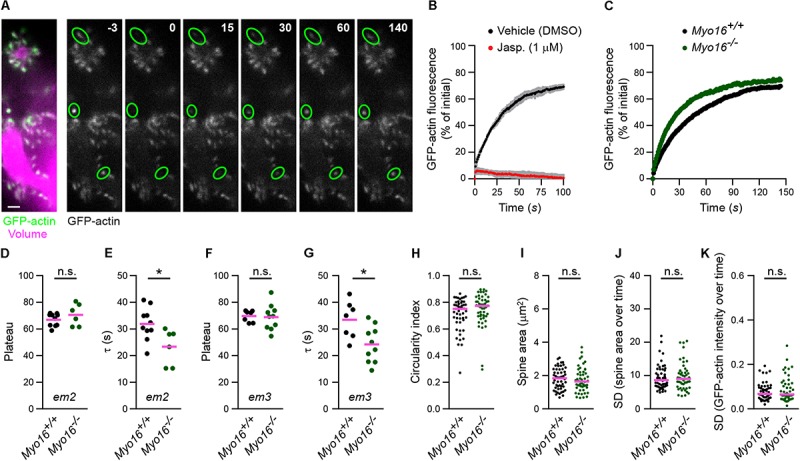
Myo16 knockout leads to accelerated F-actin turnover in Purkinje cell dendritic spines. (A) FRAP analysis of GFP-actin in PC spines. Left, Dendrite branches of a live PC at 14 DIV, co-transfected with L7/Pcp-2 promoter plasmids encoding GFP-actin (green) and volume marker FusionRed (magenta). Shown is an image recorded using spinning disk confocal microscopy. Scale bar, 2 μm. Right, Example of a FRAP experiment. Images of GFP-actin taken before and after bleaching are shown, time is indicated (seconds). Green ovals highlight bleached spines. Images correspond to frames of a time-lapse movie (Supplementary Movie S4). (B) FRAP analysis of GFP-actin in spines of PCs treated with 1 μM jasplakinolide (Jasp.; red) or with vehicle (0.1% [v/v] DMSO; black). Cells were co-transfected with L7/Pcp-2 promoter plasmids encoding GFP-actin and volume marker as in (A). Graph depicts recovery of GFP-actin fluorescence intensity in spines over time (s) relative to the bleached fluorescence intensity. Data points represent mean (n = 3 independent experiments per condition) ± SD (gray). (C) FRAP analysis of GFP-actin in spines of Myo16em2 knockout PCs (Myo16–/–) and Myo16+/+ littermate PCs co-transfected as in (A). Graph depicts recovery of GFP-actin fluorescence intensity in spines, data points represent the mean of a representative experiment (see section “Materials and Methods”). (D) GFP-actin FRAP recovery plateau in spines of Myo16em2 knockout PCs (Myo16–/–) and Myo16+/+ littermate PCs. Data show plateau values obtained as described in section “Materials and Methods” from independent experiments (n = 6–10; magenta line indicates mean); p value determined using Student’s t-test. (E) GFP-actin FRAP recovery time constant (τ) in spines of Myo16em2 knockout PCs (Myo16–/–) and Myo16+/+ littermate PCs. Data are τ values obtained as described in section “Materials and Methods” from independent experiments (n = 6–10; magenta line indicates mean); p value determined using Student’s t-test. (F,G) As in (D,E), but using Myo16em3 knockout PCs (Myo16–/–) and Myo16+/+ littermate PCs (n = 7–10). (H) Circularity index of spines of Myo16em2 knockout PCs (Myo16–/–) and Myo16+/+ littermate PCs expressing GFP-actin and volume marker. Value of 1.0 corresponds to perfectly circular shape, lower values indicate elongated shape. Data points represent single spines, magenta line indicates median; p value determined using Mann–Whitney test. (I) Apparent area covered by single spines of Myo16em2 knockout PCs (Myo16–/–) and Myo16+/+ littermate PCs expressing GFP-actin and volume marker. Data points represent single spines, magenta line indicates median; p value determined using Student’s t-test. (J) Spine area changes over time of Myo16em2 knockout PCs (Myo16–/–) and Myo16+/+ littermate PCs expressing GFP-actin and volume marker. Data points represent standard deviation (SD) of the relative area change of single spines over 150 s, magenta line indicates median; p value determined using Mann–Whitney test. (K) Change of GFP-actin fluorescence intensity over time in spines of Myo16em2 knockout PCs (Myo16–/–) and Myo16+/+ littermate PCs expressing GFP-actin and volume marker. Data points represent SD of the relative fluorescence change of single spines over 150 s, magenta line indicates median. For reason of comparability with the other figures, a single data point of Myo16–/– lying above the Y-axis limit is not shown; p value determined using Mann–Whitney test. *p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ****p < 0.0001; n.s., not significant.
