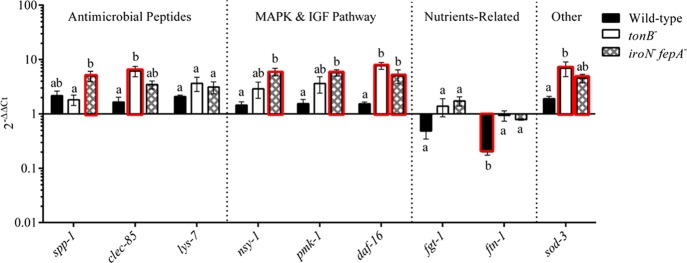
Figure 8.
Host response of C. elegans at the gene expression level to the infection of Salmonella Typhimurium wild type and mutants. Several genes related to antimicrobial peptide production, MAPK and IGF signaling pathways, and other molecules related to nutrient metabolism and defense in C. elegans were selected as the targets for the qPCR assay. The nematode was sampled on day 5 of the lifespan assay. Relative expression was determined using the 2–ΔΔCt method. The ΔCt was the comparison in the threshold cycle between target genes and housekeeping genes (snb and act-1). The ΔΔCt represented the comparison between Salmonella-infected C. elegans and C. elegans treated with E. coli OP50 only. The reference (=1) for the comparison was the gene expression in uninfected C. elegans. The RNA samples of each treatment had three biological replicates in the qPCR assay; n = 3. Means marked with “a” and “b” and those without a common letter were significantly different (P < 0.05) for the same gene among different strains. Bars with the red edge indicate a significant difference in the gene expression between infected and uninfected Caco-2 cells. tonB–, mutant defective in tonB; iroN– fepA–, mutant defective in both iroN and fepA; sod-3, superoxide dismutase 3; fgt-1, facilitated glucose transporter protein 1; ftn-1, ferritin; spp-1, saposin-like protein; clec-85, c-type lectin; lys-7, lysozyme-like protein 7; daf-16, forkhead-type transcription factor; pmk-1, mitogen-activated protein kinase pmk-1; nsy-1, mitogen-activated protein kinase kinase kinase nsy-1; IGF, insulin-like growth factor; MAPK, mitogen-activated protein kinases.