Figure 6.
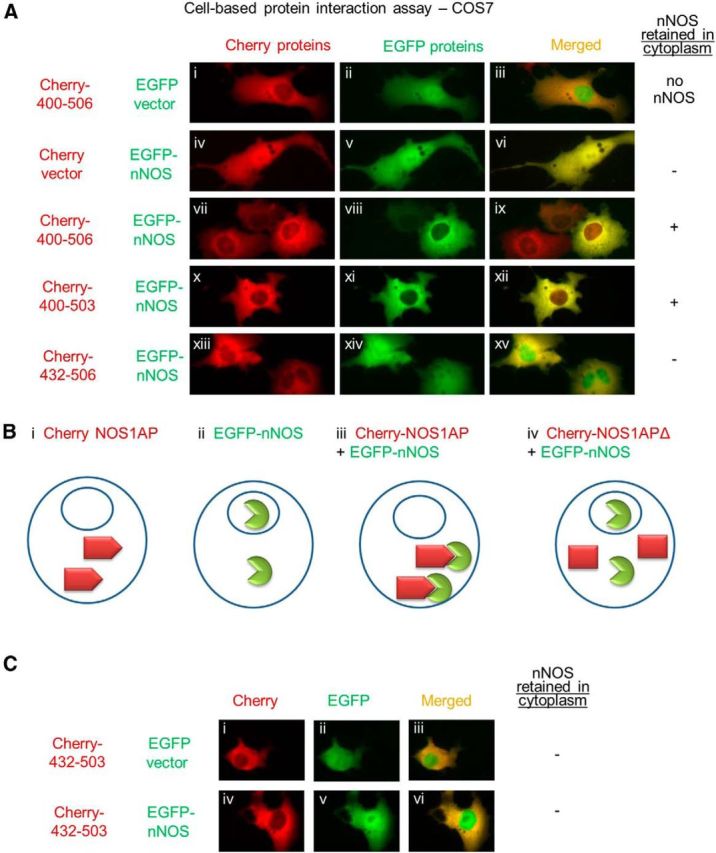
NOS1AP translocates nNOS in intact cells in a manner dependent on residues 400–431 but not the PDZ motif 504–506. A, COS7 cells were transfected with Cherry and EGFP vectors or plasmids encoding Cherry fusions with NOS1AP sequences (400–506, 400–503, 432–506) and EGFP fusions with nNOS 1–155 as indicated. B, Schematic model depicting the behavior of proteins used. Bi, All C-terminal fragments of NOS1AP used, in fusion with Cherry, reside predominantly outside the nucleus. Bii, EGFP-nNOS 1–155 enters both cytoplasmic and nuclear compartments. Biii, When both are expressed, NOS1AP localization is unchanged, but nNOS 1–155 is largely excluded from the nuclei, suggesting the formation of nNOS-NOS1AP complexes that, like NOS1AP, do not freely enter the nucleus. Biv, Deleting regions of NOS1AP that disrupt nNOS interaction is predicted to allow nNOS to enter the nucleus. Thus, deleting NOS1AP residues 400–431 eliminates the effect of NOS1AP on nNOS relocalization, whereas NOS1AP lacking the PDZ motif, residues 504–506, retains nNOS relocalization. C, Experiments were performed as in A using NOS1AP residues 432–503, lacking both PDZ motif 504–506 and the 400–432 region that appears to be required for interaction with nNOS, fused to Cherry fluorescent protein and EGFP vector (Ci–Ciii) or EGFP-nNOS1–155 (Civ–Cvi). Cherry-NOS1AP 432–503 resided predominantly in cytoplasm (Ci, Civ), but EGFP-nNOS was present in both nuclear and cytoplasmic compartments (Cv, Cvi).
